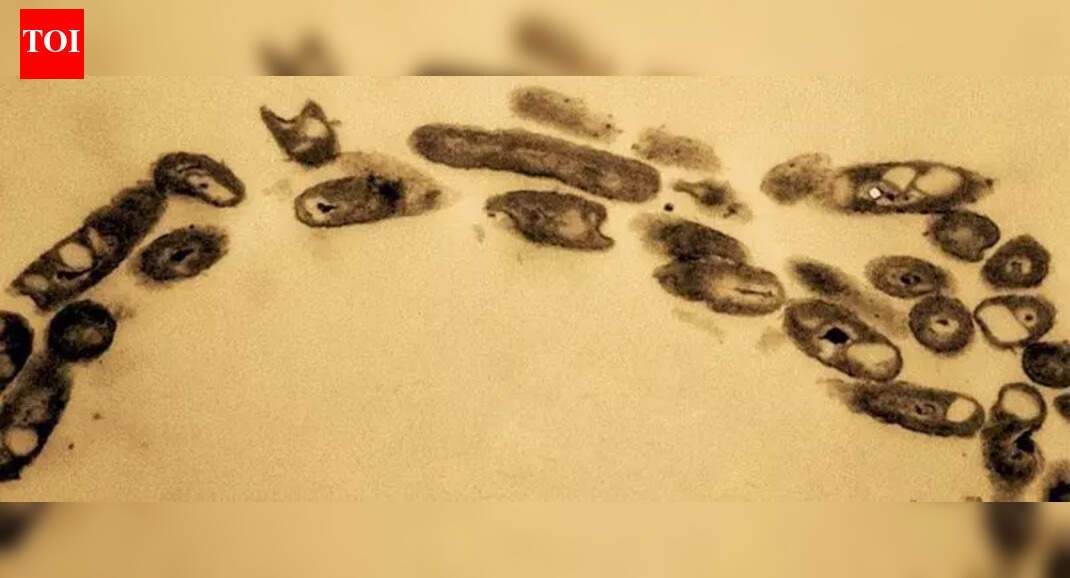
Hyderabad: Researchers who earlier identified a petroleum-eating bacterium in Nacharam have now decoded its entire genome, strengthening the possibility of using it to naturally clean up oil-polluted land.About six months ago, the research team reported isolating a new strain, Rhodococcus indonesiensis SARSHI1 from contaminated soil near petrol stations and industrial units in Nacharam. Their first study, published in Nature Scientific Reports on May 17, was based on extensive sampling using soil probes at depths between 10 cm and 1 metre.From 72 samples, five strains showed strong hydrocarbon-degrading ability and produced biosurfactants, which are natural compounds that help break down oil. Identification was done through rRNA gene sequencing and biochemical tests. Among all strains, SARSHI1 stood out for achieving nearly 90% hydrocarbon degradation while also producing high biosurfactant levels.Phytotoxicity tests showed that the bacterium is safe for the environment and adapts well to polluted soil. The researchers said this makes the strain suitable for large-scale use under industrial conditions, where it could break down petroleum hydrocarbons while simultaneously generating biosurfactants. The study placed these findings against the wider backdrop of oil pollution caused by spills and industrial discharge.What is it capable of? In the latest study, published on Dec 2 in Scientific Reports, researchers led by Anuraj Nayarisseri (In Silico research laboratory, eminent biosciences, Indore) and Rajabrata Bhuyan (department of bioscience and niotechnology, Banasthali Vidyapith, Rajasthan) presented a complete genome map of the Nacharam strain.Using a hybrid sequencing method combining Oxford Nanopore Technologies and Illumina, they assembled a 5.7 megabase pairs (Mbp) genome along with a 159,118 base pairs (bp) plasmid. Together, these carry 5,150 coding sequences. Structural annotation revealed 5,220 genes. The genome showed 100% completeness with a coding density of 91.4%.Linked to oil breakdownFunctional analysis identified key genes responsible for degrading long-chain alkanes (alkB, ahyA, almA) and aromatic hydrocarbons (bph, ben, xylC). The strain also carries genes for multiple antibiotic resistance and secondary metabolite production, highlighting its adaptability.The researchers said this strong genetic toolkit, supported by earlier field tests and safety studies, shows that R. indonesiensis SARSHI1 could be used as an effective, natural solution for cleaning petroleum-polluted land. The genome data is now available in GenBank.