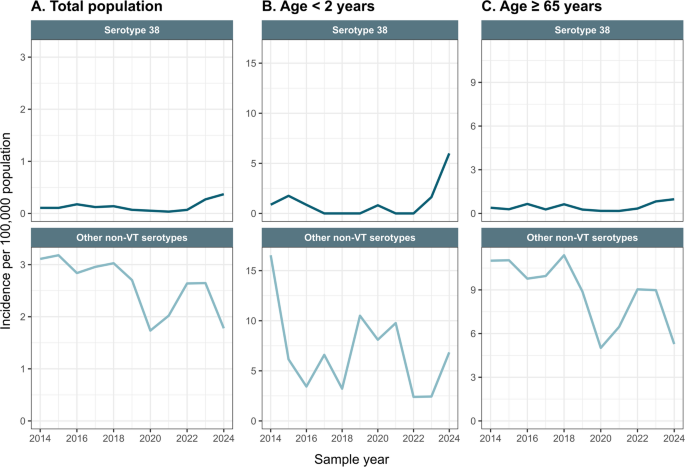
IPD was defined as isolating S. pneumoniae by culture from a normally sterile site, e.g., blood or cerebrospinal fluid. We included all IPD cases from Danish residents with a serotype not included in PCV13 or PPV23, hereafter referred to as non-VT. Isolates, where the serotype could only partially be determined (e.g. group 12) and where one of the serotypes in the group (e.g. serotype 12F) is included in either PCV13 or PPV23, were excluded. See supplementary table S1 for identified non-VT serotypes included. Serotype 38 IPD was compared to all other non-VT IPD cases during the period. Annual incidence rates per 100,000 population and 95% confidence interval (CI) were calculated for each group from 2014 to 2024, using population data from Statistics Denmark8. For clinical characterization, antimicrobial susceptibility and whole-genome sequencing (WGS) of serotype 38 IPD, we limited the period to include cases from January 2022 to September 2024, because the rise in serotype 38 IPD began in 2023.
Molecular characteristics
Since 2007, sending pneumococcal isolates from sterile sites to Statens Serum Institut (SSI) for characterization has been mandatory. Since then, all IPD isolates have been phenotyped as described by Golden et al.9. WGS was performed on all serotype 38 IPD isolates as previously described, using the Bifrost pipeline or quality control10,11,12. Multilocus sequence typing (MLST) for each isolate was conducted using the Bifrost pipeline, linked to the S. pneumoniae database at PubMLST13. Genotyping was performed using PneumoCaT (v1.2.1)11,14.
Phylogenetic analysis
A phylogenetic tree was created based on single-nucleotide polymorphisms (SNPs) detected in the core genome of the isolate collection based on the major PGSC lineages. SNPs were identified using NASP15 with BWAmem for mapping against the chromosome of S. pneumoniae isolate, and GATK was set to remove positions with less than 10-fold depth and 90% unambiguous variant calls after removal of duplicated regions in the reference using NUCmer. The resulting SNP matrix was purged for recombination using Gubbins16. The genomic sequence data for the 42 serotype 38 clinical isolates have been deposited at the European Nucleotide Archive (ENA) under Bioproject no. PRJEB88491. The phylogenetic tree was generated using CLC Main Workbench (Version 22.0.2).
Antimicrobial susceptibility
All isolates of S. pneumoniae sent to SSI is tested for antibiotic susceptibility performed as previously described17. Screening was done according to EUCAST (www.eucast.org) by disk diffusion using Mueller–Hinton 5% blood agar with NAD (Oxoid, Denmark) incubated in ambient air with 5% CO2 at 35 °C and oxacillin, erythromycin and clindamycin discs (Oxoid, Denmark). Isolates showing non-susceptibility (oxacillin resistant if zone diameter
Clinical characteristics
Baseline characteristics included age, clinical focus, and underlying medical conditions. Hospital admissions for IPD were defined as sampling occurring during admission or within five days prior, provided the admission was related to IPD (supplementary table S2). All IPD-related admissions were included. Length of stay was calculated from admission to discharge date or from sampling to discharge date if sampling date occurred more than two days after admission. Data on underlying medical conditions, length of hospital stay, intensive care unit treatment, and 30-day case fatality were retrieved from the Danish National Patient Register and the Danish Civil Register. The International Classification of Diseases 10th Revision (ICD-10) codes18 and the Danish Health Care Classification System (Sundheds-væsenets Klassifikations System, SKS) procedure codes19 were used to identify underlying medical conditions, IPD-related admissions and intensive care treatment (supplementary table S2). Vaccination status for all children under the age of two years was retrieved from the Danish Vaccination Register3.
Statistical analysis
Comparisons between groups were performed using Chi-square test for categorical data and Kruskal-Wallis test for continuous variables with statistical significance defined as a p-value 20.
Ethics approval
SSI processes personal data under the authority of Article 6(1)(e) of the General Data Protection Regulation (GDPR), in conjunction with Sect. 222 of the Danish Health Act, as the processing is necessary for the performance of a task assigned to SSI. As part of its monitoring duties, SSI manages the national systems for reportable diseases, this including IPD and since data were collected routinely for national surveillance purposes no ethical approval or informed consent was required. Data from national administrative registers were linked with the IPD cases and according to Danish law, ethical approval is not needed for this type of research. All methods were carried out in accordance with relevant guidelines and regulations.