Most AI pipelines throw away structure and meaning to compress data.
I built something that doesn’t.
What I Built: A Lossless, Structure-Preserving Matrix Intelligence Engine
Use it to:
- Find connections between datasets (e.g., drugs ↔ genes ↔ categories)
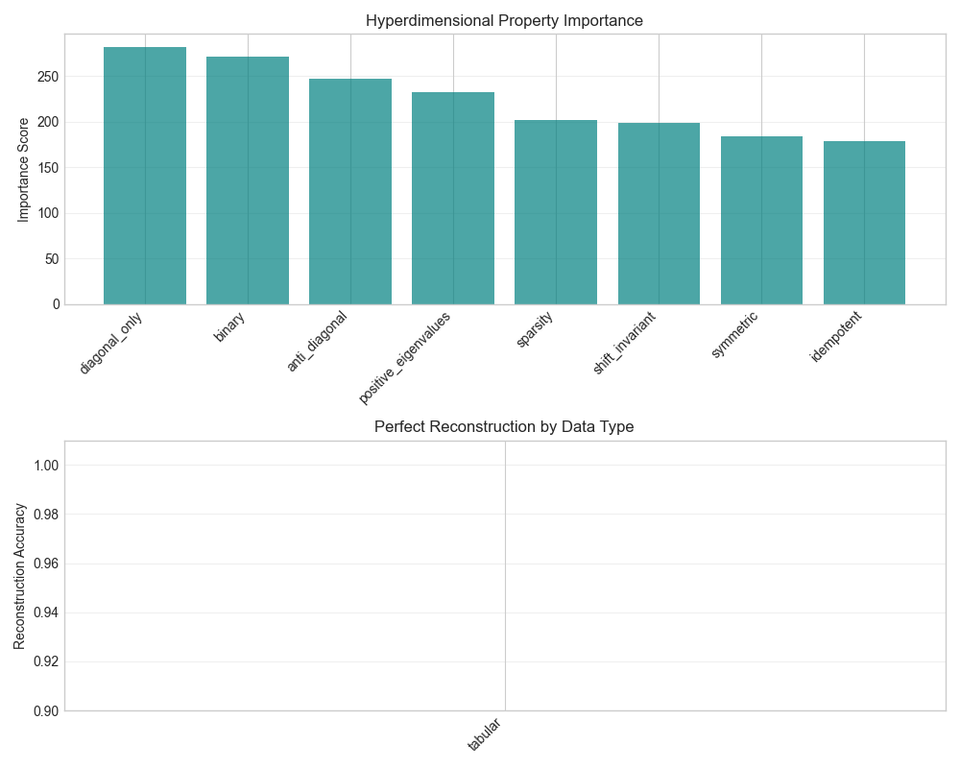
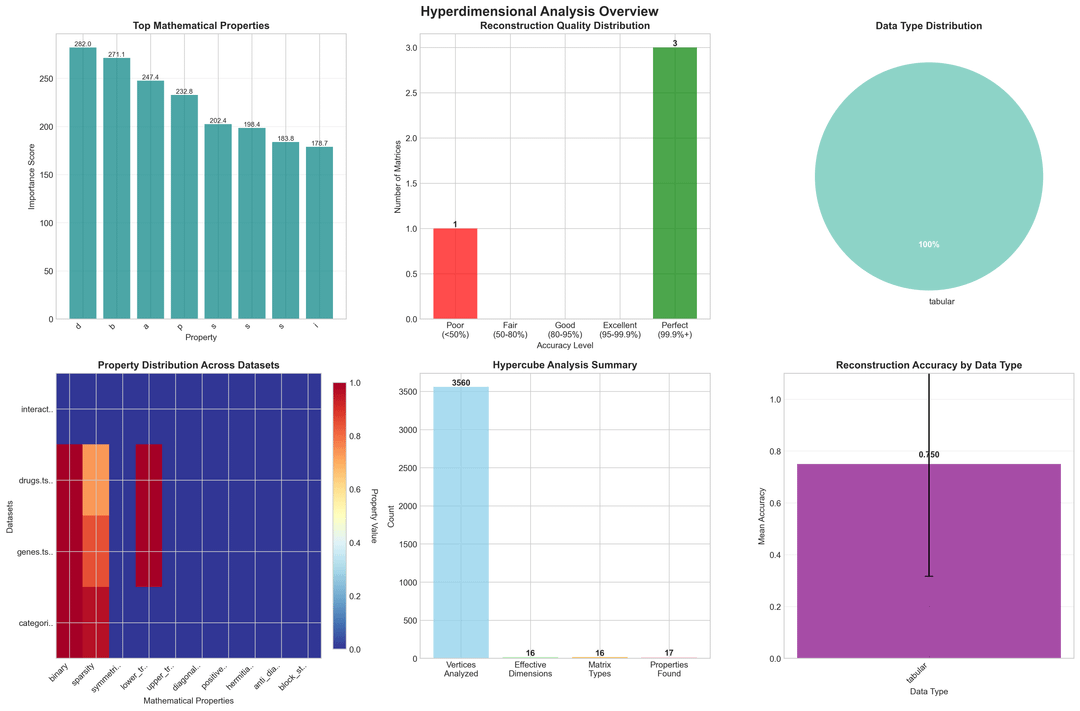
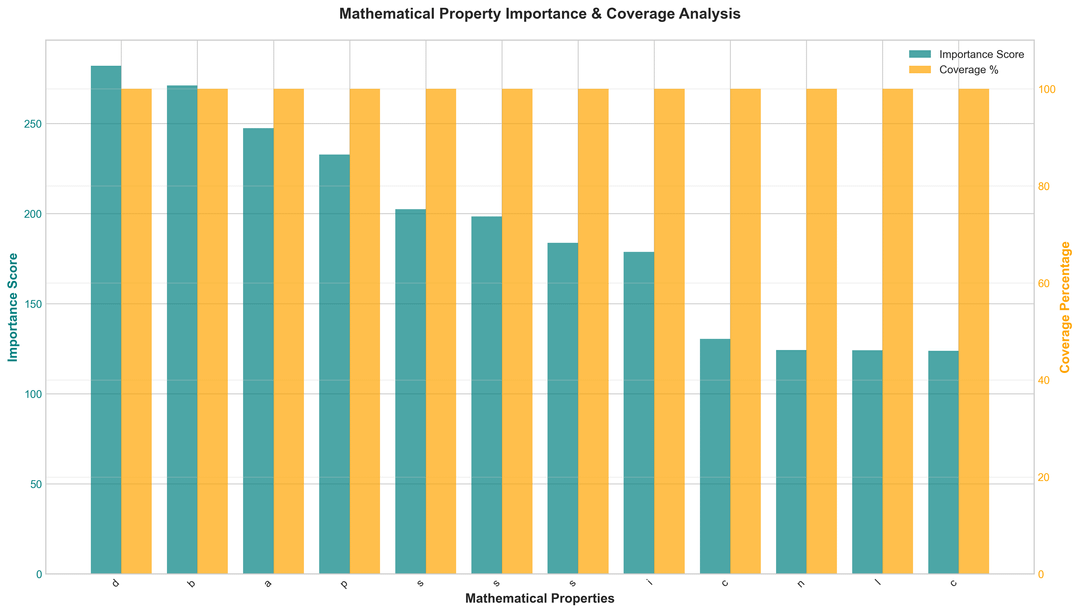
- Analyze matrix structure (sparsity, binary, diagonal)
- Cluster semantically similar datasets
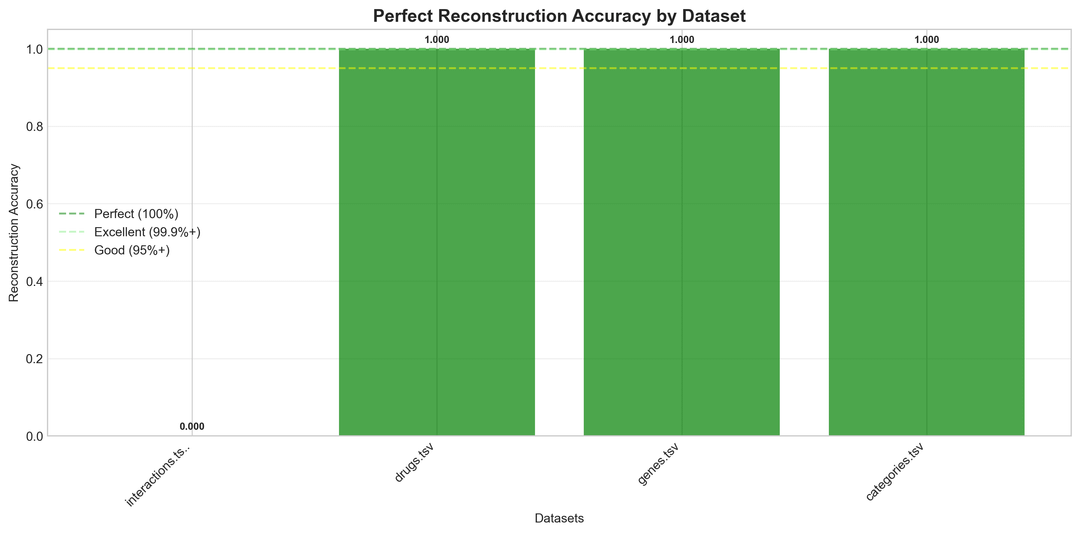
- Benchmark reconstruction (up to 100% accuracy)
No AI guessing — just explainable structure-preserving math.
Key Benchmarks (Real Biomedical Data)
Try It Instantly (Docker Only)
Just run this — no setup required:
bashCopyEditmkdir data results
# Drop your TSV/CSV files into the data folder
docker run -it \
-v $(pwd)/data:/app/data \
-v $(pwd)/results:/app/results \
fikayomiayodele/hyperdimensional-connection
Your results show up in the results/folder.
Installation, Usage & Documentation
All installation instructions and usage examples are in the GitHub README:
📘 github.com/fikayoAy/MatrixTransformer
No Python dependencies needed — just Docker.
Runs on Linux, macOS, Windows, or GitHub Codespaces for browser-only users.
📄 Scientific Paper
This project is based on the research papers:
Ayodele, F. (2025). Hyperdimensional connection method – A Lossless Framework Preserving Meaning, Structure, and Semantic Relationships across Modalities.(A MatrixTransformer subsidiary). Zenodo. https://doi.org/10.5281/zenodo.16051260
Ayodele, F. (2025). MatrixTransformer. Zenodo. https://doi.org/10.5281/zenodo.15928158
It includes full benchmarks, architecture, theory, and reproducibility claims.
🧬 Use Cases
- Drug Discovery: Build knowledge graphs from drug–gene–category data
- ML Pipelines: Select algorithms based on matrix structure
- ETL QA: Flag isolated or corrupted files instantly
- Semantic Clustering: Without any training
- Bio/NLP/Vision Data: Works on anything matrix-like
💡 Why This Is Different
| Feature | Traditional Tools | This Tool |
|---|---|---|
| Deep learning required | ✅ | ❌ (deterministic math) |
| Semantic relationships | ❌ | ✅ 99.999%+ similarity |
| Cross-domain support | ❌ | ✅ (bio, text, visual) |
| 100% reproducible | ❌ | ✅ (same results every time) |
| Zero setup | ❌ | ✅ Docker-only |
🤝 Join In or Build On It
If you find it useful:
- 🌟 Star the repo
- 🔁 Fork or extend it
- 📎 Cite the paper in your own work
- 💬 Drop feedback or ideas—I’m exploring time-series & vision next
This is open source, open science, and meant to empower others.
📦 Docker Hub: fikayomiayodele/hyperdimensional-connection
🧠 GitHub: github.com/fikayoAy/MatrixTransformer
Looking forward to feedback from researchers, skeptics, and builders
Posted by Hyper_graph
3 comments
Idk if it’s just me reading in bed too late but nothing in the “paper” or this post makes any sense at all lol
I think you gotta lay off the chatgpt
I’m a little confused by the papers you’ve submitted. This isn’t my field at all, but I *do* have a number of years of experience in academia so I’ve ended up with some questions:
* The Zenodo papers haven’t been peer-reviewed. Have you submitted them to any peer-reviewed journals?
* Is the raw data from either paper available? I could not see it.
* Why have you referenced no other authors than yourself? This is unheard-of in genuine research.
* You’ve clearly put some effort into the appearance of your paper, but your references section doesn’t use a consistent referencing convention. Why is this?
It’d take an expert in the field to really assess the software itself, but these appear to be red flags on a general level.
And the plots are either empty or flat
Comments are closed.