According to WHO9, ‘infectious respiratory particles’ (IRPs) are potentially infectious particles generated by individuals infected with a respiratory pathogen, during the infectious stage of the disease. The pathogen containing IRPs, along with water and respiratory secretions, carried by expired airflow, exit the infectious person’s mouth/nose through breathing, talking, singing, spitting, coughing or sneezing and enter the surrounding air. Besides IRPs, water and electrolytes, mucosalivary fluid may also contain various cell types (e.g., epithelial cells and cells of the immune system)35. Contaminated surfaces are thus created when IRPs expelled into the air settle on a surface, or when an infected person transfers infectious respiratory secretions by firstly touching their own mouth, nose or eyes and then touching a surface36. Infectious pathogens on the contaminated surfaces are then transferred to another person who touches that contaminated surface and then their own mouth, nose or eyes. This is commonly known as indirect contact transmission.
Environmental monitoring of respiratory viruses is thus essential for preventing and containing epidemics, although their detection at very low concentrations could sometimes be challenging. This kind of surveillance, if routinely performed, especially in seasonal periods in which respiratory virus circulation is of particular concern for public health, could be a useful tool to promptly recognise an increase of their presence and to identify spaces in which sanitation measures could be particularly needed.
Several authors37,38,39,40,41, including our research group26 recently described the high sensitivity offered by the application of molecular-based systems for the environmental detection of SARS-CoV-2 or other airborne pathogens in crowded public buildings. Molecular detection allows to shorten detection time, increase sensitivity, and circumvent drawbacks associated with conventional viral culture42. However, the reliability of molecular detection methods in environmental epidemiological surveillance is mainly based on proper assessment and correct use of process controls, especially if considering the well-known inhibitory effect of contaminants of environmental origin on the amplification reactions.
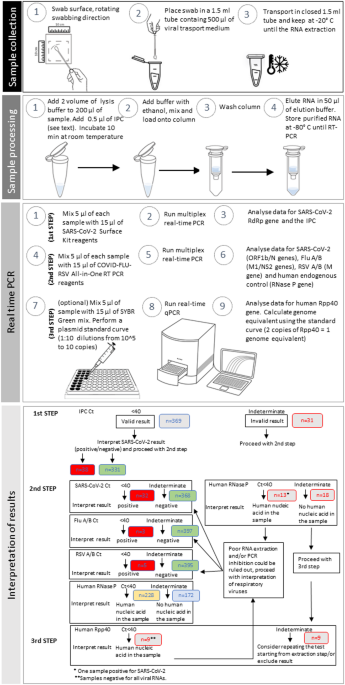
In this study, a new Surface 3-step PCR platform, introducing a novel approach suitable to assess result reliability, has been developed. This system included a first phase in which an Internal Process Control (IPC), a synthetic RNA oligonucleotide, was added to the environmental sample during lysis, to be co-extracted and co-amplified along with the viral target. Thus, the presence/absence of the IPC-specific amplification product allowed to evaluate RNA extraction efficiency and identify the possible presence of PCR inhibitors. In our work, 31/400 (7.8%) samples gave an invalid test result (IPC Ct > 40), probably due to low recovery in the nucleic acid extraction/isolation procedure or the presence of PCR inhibitors or a low PCR efficiency of IPC amplification. Concerning potential inhibitors in this kind of samples, the presence of detergents used for surface cleaning could be hypothesized. Additionally, the inhibitory effect of the sampling swab itself, depending on its composition, has been previously described43. The use of a sampling/transport medium containing suitable neutralizing agents could be a potential solution. However, the first step of the method applied in our work allowed to detect SARS-CoV-2 RNA in 9.5% of samples, with a decreasing trend during time intervals and a main prevalence on student desks.
The second step of our approach provided results about a panel of three main respiratory viruses (SARS-CoV-2, influenza virus Flu A/B and respiratory syncytial virus RSV A/B), along with human RNase P gene as endogenous control. Indeed, although the human RNase P gene is not strictly an endogenous control for such environmental samples, it demonstrates the presence or passage of people in the sampling locations and the elimination of human DNA vehiculated by mucosalivary fluids. Moreover, the usefulness of exogenous control is restricted to the monitoring of possible errors in nucleic acid extraction and amplification, but not during sampling and pre-PCR handling. In contrast, the importance of the RNase P target relies on its natural presence in the target environment, offering the opportunity to monitor the entire workflow and avoiding further sample manipulation44. In our study, the proper amplification of RNaseP allowed to include in the analysis additional 13 samples, which were previously assigned as invalid in step 1.
Our results highlighted a certain rate of disagreement between samples testing positive for SARS-CoV-2. The detection of SARS-CoV-2 RNA, based on different target sequences with respect to that of step 1 (ORF1b/N vs. RdRp), allowed to confirm the positivity of 16 samples, to detect 16 additional samples, but failed to identify 22 samples which tested positive in the first phase. A recent comparison highlighted that assays targeting the N gene were more specific than those targeting the RdRp45. Various real time RT-PCR assays are currently used worldwide, targeting different genes of the SARS-CoV-2, such as the envelope (E), ORF1ab/RNA-dependent RNA polymerase (RdRp), nucleocapsid (N), and spike (S) genes46. However, different RT-PCR assays would not be expected to have similar sensitivity, even when testing the same sample and several factors can affect the final Ct value such as thermocycling parameters and the optical threshold of detection47. As reported by Jeong et al.48, sensitivity and/or specificity of different PCR-based methods are dependent on genes and primer/probes selected. Although the primers and probes targeted conserved regions of the viral genome sequences, the variation of SARS-CoV-2 RNA sequence can produce mismatches in binding, leading to differences in the assay performance and potential false negative results. For this reason, a combination of targets is generally preferred for enhanced reliability.
However, taking into account both targets used in this study, a total of 54 SARS-CoV-2 positive samples, corresponding to 13.5%, were retrieved. Our discordant results cannot be ascribed to the uneven distribution of target RNA on surfaces, since the two amplification kits have been used on the same extracted sample. On the contrary, although samples have been correctly stored at −80 °C, the potential degradation of the viral RNA, especially at the very low concentration found in our samples, cannot be excluded. Finally, concerning the influence of matrix-associated inhibitors, it should be noted that the first kit of the multistep assay has been previously tested by us on different materials (plastic, metal, wood and paper) and the results reported in Casabianca et al.26 indicated that none of the four specific matrices affected RNA isolation, SARS-CoV-2 RNA detection or inhibited PCR, as revealed by the positive amplification signals of both the IPC and RdRp gene, reported in the above-mentioned article. The key element to explain the high number of discordant results should be sought, instead, in the very low amount of viral genomes in the environmental surfaces. This consideration is supported by Eibinger et al.49, who reported that SARS-CoV-2 RT-qPCR assays may show considerable variability, especially in samples with low viral RNA concentrations. FluA/B and RSV A/B were also detected with very low prevalence in the second and third time-periods, in line with their common seasonal distribution5.
Positive samples were mainly obtained both from classrooms in which students attended their lessons and from studying and dining tables, sampled immediately after lesson/lunch end and student exit. In contrast, samples from common areas (i.e., toilets, vending machine push-button panels) were always negative. These results could be explained by the fact that common areas’ cleaning and sanitizing is made more frequently than classrooms’, that are closed during lesson hours. Moreover, students and teachers reside longer in classrooms than in common areas, that are located in wide corridors and in places of passage, and the actual probability of contamination will increase with longer duration of staying in a room. Also, crowding level is different among those locations.
Finally, the third phase of the method consisted in the quantitative detection of hRpp40, a further single copy endogenous control for human DNA. This last step allowed the detection of human DNA in 9 additional samples, which were assigned as invalid in step 2. Thanks to the combined multistep analysis, the number of invalid samples was considerably reduced and the examination of the 400 samples led to a significant performance improvement in detecting viral RNA on environmental surfaces, going from 1 failed sample out of 13 to 1 out of 44, with a very high prevalence of valid results (98%).
The overall prevalence of SARS-CoV-2 in university surfaces reported by other authors is quite variable. Zhang et al.40, who carried out an investigation on a university campus located in the U.S. in 2020–2021, found a 1.4% prevalence of SARS-CoV-2 positive surface samples. Interestingly, these authors proved that the total case number on campus was significantly higher in weeks with positive environmental samples than in non-positive weeks (Point-biserial correlation, p = 0.001). Another study, conducted in 2023 at a university in Southern Italy50, reported a slightly higher prevalence of 5.4%, with a direct association between the average number of COVID-19 cases among university students in the seven days following sampling and the percentage of SARS-CoV-2 positive swabs on sampling day. A similar prevalence was detected in a previous study of our research group, carried out in Autumn-Winter 2021 in Central Italy26. However, a relevant point reported by De Giglio et al.50 was the greater contamination percentage found in the Spring–Summer period than in the Autumn, suggesting that an effective monitoring plan, to be implemented as an early warning system, could benefit from a more extensive sampling timespan. Finally, in the interval January-November 2022, Ali et al.39, sampling environmental surfaces in a large university campus in Florida, reported a very high SARS-CoV-2 viral RNA presence (90.7%), correlating with trends in community-level activity and case reports from the student health centre. In the present study, a percentage of positive samples corresponding to 16% resulted positive for the RNA of all the respiratory viruses tested, of which 14% for the sole SARS-CoV-2, only in the time interval late Autumn-early Winter. A comparison among the epidemiological data reported by different authors is nonetheless difficult, since data are strongly dependent on different factors, such as the epidemiological situation, virus circulation (including the various types and variants), mandatory use of personal protective equipment or other restrictions, along with the specific detection protocol used for surface sampling, extraction and amplification. However, since several students come daily or weekly from other cities as commuters, public data on the prevalence of diseases caused by these three pathogens in the University town could not mirror the epidemiological situation present at the Campus.
As outlined elsewhere40, the university environment represents a nonhealthcare but potentially risky setting, especially if considering lecture halls and exercise facilities51, in which the crowding or the peculiar activity performed, or also the sharing of tables and devices/equipment (i.e., PCs) determine a higher transmission potential. The model for quantitative microbial risk assessment, applied to estimate the probability of infection after exposure to SARS-CoV-2 by contact with contaminated surfaces, revealed a correlation between COVID-19 case numbers and the environmental positive samples39,40,50. Unfortunately, in our study the number of cases among the university population was not determined, but a similar correlation could be reasonably hypothesized and extended also to other respiratory viruses, such as those detected in this investigation. Therefore, the monitoring of surface environmental samples may represent a convenient indicator of potential infection spread in the community. The information obtained from high-touch surface monitoring could also allow to identify hotspot areas, requiring more frequent cleaning, sanitation and ventilation. Although the survival of viruses on surfaces is influenced by numerous factors, and the actual probability of transmission leading to infection via this route (fomite transmission) is likely not the primary one, the purpose of the environmental surveillance in this university setting had the primary objective of determining an early warning system to assess and mitigate the potential transmission risk in the specific environments in which samplings have been carried out. Indeed, the presence of viral genetic material on the surfaces reveals the transit and contact of infected individuals. Further objectives included the identification of potential transmission hotspots as priority areas for targeted intervention (i.e., sanitation) and the recognition of critical timeframes for enhanced prevention. Even if surface testing is a complement to preventive measures and environmental monitoring plans, it remains useful as part of the risk assessment to also ensure student and employees’ safety.
Results of this study allowed us to formulate specific prevention and control recommendations. We provided some evidence to suggest that there is an effect of enhancing class size to reduce the chance of finding positive samples. Small classrooms and inside them, student desks and teacher workstations, have been identified as priority areas for targeted interventions, such as daily cleaning and sanitation. Further specific measures may include the use of air sanitation devices providing microbial inactivation52. Moreover, at least in our investigation, the period of late Autumn (weeks 48–49) and the beginning of the new year (weeks 2–4) has been recognised as a critical timeframe, during which the application of the above-mentioned measures could offer the benefit of enhanced prevention. Moreover, the increment in Ct values recorded over the three time periods suggests a decrement in viral RNAs. The complexity of the academic environment, along with the several factors impacting on viral presence on surfaces (viral load, material, location, crowding, number of infected people, sanitation products, etc.), may influence sample recovery and test results. Therefore, proper method selection is a key element in environmental surveillance.
Our approach may represent a promising tool for non-healthcare public place as a university setting in surveilling fomites associated with respiratory viruses or other pathogen’s transmission. Although the 3-Step method is complex for routine monitoring, in this study, the combined use of the three assays allowed us to understand their usefulness and limitations, particularly in the detection of false negatives. Indeed, the complementary use of the 3 QCs allowed us to monitor the efficiency of the RNA extraction procedure and the effectiveness of the PCR reaction, and thus exclude the presence of false negatives with a sensitivity of 98%. However, testing all samples first, then performing QC only on the negative samples, could potentially reduce the number of steps and overall workload.
Finally, our goal was not to compare kits but to maximize detection while minimizing the number of invalid samples. The IPC, being an exogenous synthetic RNA fragment, may be unstable and consequently become un-amplifiable. On the contrary, an endogenous control (such as the endogenous human RNaseP of the second step method), is more reliable in revealing nucleic acid amplifiability. Finally, the introduction of a further endogenous target (Rpp40 endogenous control) offers the possibility to amplify a different human DNA fragment33, with possibly different amplification efficiency, thus complementing and integrating the so-called endogenous control system. The use of multiple (at least two) target viral sequences (i.e., RdRp and ORF plus N) is always highly recommended53 since some genetic fragments may contain sequence mutations that can affect detection reliability.
The operating protocol, which we describe in this work and extensively tested previously26, can be easily implemented using an automatic nucleic acid extractor. For example, the one present in our laboratories allows the extraction of 32 samples in 10–15 min with only a few manual steps by the operator, utilizing magnetic bead-based technology. Three nucleic acid extraction sessions (96 samples) can then be analyzed all together in a single real-time PCR plate (~ 2 h). Therefore, the analysis of approximately 100 samples can be done in one working day.
This medium-throughput protocol (extraction with a small automated extractor and semi-automated real-time PCR) does not require large spaces and sophisticated environmental surveillance laboratories.
Limitations of our study include the lack of correlation with epidemiologic data, the minimal detection of viruses other than SARS-CoV-2 and the absence of validation data due to the use of commercial assays with analytical sensitivity determined by synthetic RNA controls rather than environmental matrices.