Fiest, K. M. et al. Prevalence and incidence of epilepsy: a systematic review and meta-analysis of international studies. Neurology 88, 296–303 (2017).
Begley, C. E. & Beghi, E. The economic cost of epilepsy: a review of the literature. Epilepsia 43, 3–9 (2002).
Tavakol, S. et al. Neuroimaging and connectomics of drug-resistant epilepsy at multiple scales: from focal lesions to macroscale networks. Epilepsia 60, 593–604 (2019).
Fiest, K. M., Birbeck, G. L., Jacoby, A. & Jette, N. Stigma in epilepsy. Curr. Neurol. Neurosci. Rep. 14, 444 (2014).
Tellez-Zenteno, J. F., Patten, S. B., Jetté, N., Williams, J. & Wiebe, S. Psychiatric comorbidity in epilepsy: a population-based analysis. Epilepsia 48, 2336–2344 (2007).
Sultana, B. et al. Incidence and prevalence of drug-resistant epilepsy: a systematic review and meta-analysis. Neurology 96, 805–817 (2021).
Kwan, P. et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia 51, 1069–1077 (2010).
Engel, J. What can we do for people with drug-resistant epilepsy? The 2016 Wartenberg Lecture. Neurology 87, 2483–2489 (2016).
Perucca, P., Bahlo, M. & Berkovic, S. F. The genetics of epilepsy. Annu. Rev. Genom. Hum. Genet. 21, 205–230 (2020).
Qiu, Y. et al. On-demand cell-autonomous gene therapy for brain circuit disorders. Science 378, 523–532 (2022).
International League Against Epilepsy Consortium on Complex Epilepsies. GWAS meta-analysis of over 29,000 people with epilepsy identifies 26 risk loci and subtype-specific genetic architecture. Nat Genet. 55, 1471–1482 (2023).
Li, R. et al. Transcriptionally downregulated GABAergic genes associated with synaptic density network dysfunction in temporal lobe epilepsy. Eur. J. Nucl. Med. Mol. Imaging https://doi.org/10.1007/s00259-024-07054-5 (2025).
François, L. et al. Identification of gene regulatory networks affected across drug-resistant epilepsies. Nat. Commun. 15, 2180 (2024).
Alsubhi, S. et al. Utility of genetic testing in the pre-surgical evaluation of children with drug-resistant epilepsy. J. Neurol. 271, 2503–2508 (2024).
Li, Y. et al. Alterations in spontaneous brain activity and functional network reorganization following surgery in children with medically refractory epilepsy: a resting-state functional magnetic resonance imaging study. Front. Neurol. 8, 374 (2017).
Larivière, S., Bernasconi, A., Bernasconi, N. & Bernhardt, B. C. Connectome biomarkers of drug-resistant epilepsy. Epilepsia 62, https://doi.org/10.1111/epi.16753 (2021).
Johnson, G. W., Doss, D. J. & Englot, D. J. Network dysfunction in pre and postsurgical epilepsy: connectomics as a tool and not a destination. Curr. Opin. Neurol. 35, 196–201 (2022).
Shao, R. et al. Alteration in early resting‑state functional MRI activity in comatose survivors of cardiac arrest: a prospective cohort study. Crit. Care 28, 260 (2024).
Jing, J. et al. Central vein sign and trigeminal lesions of multiple sclerosis visualised by 7T MRI. J. Neurol. Neurosurg. Psychiatry 95, 761–766 (2024).
Hagen, J. et al. Phenomena of hypo- and hyperconnectivity in basal ganglia-thalamo-cortical circuits linked to major depression: a 7T fMRI study. Mol. Psychiatry 30, 158–167 (2025).
Khoshkhoo, S. et al. Contribution of somatic Ras/Raf/mitogen-activated protein kinase variants in the hippocampus in drug-resistant mesial temporal lobe epilepsy. JAMA Neurol. 80, 578–587 (2023).
Pfisterer, U. et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 11, 5038 (2020).
Sunkin, S. M. et al. Allen brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 41, https://doi.org/10.1093/nar/gks1042 (2013).
Estevez-Fraga, C. et al. Genetic topography and cortical cell loss in Huntington’s disease link development and neurodegeneration. Brain 146, 4532–4546 (2023).
Arnatkeviciute, A., Markello, R. D., Fulcher, B. D., Misic, B. & Fornito, A. Toward best practices for imaging transcriptomics of the human brain. Biol. Psychiatry 93, 391–404 (2023).
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012).
Williams, J. A. et al. Inflammation and brain structure in schizophrenia and other neuropsychiatric disorders: a mendelian randomization study. JAMA Psychiatry 79, 498–507 (2022).
Qin, L. et al. Dynamic functional connectivity and gene expression correlates in temporal lobe epilepsy: insights from hidden Markov models. J. Transl. Med. 22, 763 (2024).
Sun, F. et al. Hippocampal gray matter volume alterations in patients with first-episode and recurrent major depressive disorder and their associations with gene profiles. BMC Psychiatry 25, 134 (2025).
Zhu, J. et al. Transcriptomic decoding of regional cortical vulnerability to major depressive disorder. Commun. Biol. 7, 960 (2024).
Knowles, J. K. et al. Precision medicine for genetic epilepsy on the horizon: recent advances, present challenges, and suggestions for continued progress. Epilepsia 63, 2461–2475 (2022).
Blokland, G. A. M., de Zubicaray, G. I., McMahon, K. L. & Wright, M. J. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res. Hum. Genet. 15, 351–371 (2012).
Arnatkeviciute, A., Fulcher, B. D. & Fornito, A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage 189, 353–367 (2019).
Li, J. et al. Cortical structural differences in major depressive disorder correlate with cell type-specific transcriptional signatures. Nat. Commun. 12, 1647 (2021).
Menon, V. 20 years of the default mode network: a review and synthesis. Neuron 111, 2469–2487 (2023).
Li, Q. et al. Linked patterns of symptoms and cognitive covariation with functional brain controllability in major depressive disorder. EBioMedicine 106, 105255 (2024).
Jiang, L. et al. Multimodal covariance network reflects individual cognitive flexibility. Int. J. Neural Syst. 34, 2450018 (2024).
Sun, C.-C. et al. Modified constraint-induced movement therapy enhances cortical plasticity in a rat model of traumatic brain injury: a resting-state functional MRI study. Neural Regen. Res. 18, 410–415 (2023).
Luo, G. et al. Abnormal ReHo and ALFF values in drug-naïve depressed patients with suicidal ideation or attempts: evidence from the REST-meta-MDD consortium. Prog. Neuropsychopharmacol. Biol. Psychiatry 136, 111210 (2024).
Qin, Y. et al. Rhythmic network modulation to thalamocortical couplings in epilepsy. Int. J. Neural Syst. 30, 2050014 (2020).
Li, R. et al. Epileptic discharge related functional connectivity within and between networks in benign epilepsy with centrotemporal spikes. Int. J. Neural Syst. 27, 1750018 (2017).
Akyuz, E., Arulsamy, A., Hasanli, S., Yilmaz, E. B. & Shaikh, M. F. Elucidating the visual phenomena in epilepsy: a mini review. Epilepsy Res. 190, 107093 (2023).
Wang, K. et al. Vagus nerve stimulation balanced disrupted default-mode network and salience network in a postsurgical epileptic patient. Neuropsychiatr. Dis. Treat. 12, 2561–2571 (2016).
Bacon, E. J. et al. Functional and effective connectivity analysis of drug-resistant epilepsy: a resting-state fMRI analysis. Front. Neurosci. 17, 1163111 (2023).
Widjaja, E., Zamyadi, M., Raybaud, C., Snead, O. C. & Smith, M. L. Impaired default mode network on resting-state FMRI in children with medically refractory epilepsy. AJNR Am. J. Neuroradiol. 34, 552–557 (2013).
Zhou, H.-X. et al. Rumination and the default mode network: meta-analysis of brain imaging studies and implications for depression. Neuroimage 206, 116287 (2020).
Zhang, Z. et al. Longitudinal assessment of resting-state fMRI in temporal lobe epilepsy: a two-year follow-up study. Epilepsy Behav. 103, 106858 (2020).
Dosenbach, N. U. F., Raichle, M. E. & Gordon, E. M. The brain’s action-mode network. Nat. Rev. Neurosci. https://doi.org/10.1038/s41583-024-00895-x (2025).
Englot, D. J. et al. Functional connectivity disturbances of the ascending reticular activating system in temporal lobe epilepsy. J. Neurol. Neurosurg. Psychiatry 88, 925–932 (2017).
Motelow, J. E. et al. Decreased subcortical cholinergic arousal in focal seizures. Neuron 85, 561–572 (2015).
Englot, D. J. et al. Remote effects of focal hippocampal seizures on the rat neocortex. J. Neurosci. 28, 9066–9081 (2008).
Sainburg, L. E. et al. Structural disconnection relates to functional changes after temporal lobe epilepsy surgery. Brain 146, 3913–3922 (2023).
Ganos, C. et al. A neural network for tics: insights from causal brain lesions and deep brain stimulation. Brain 145, 4385–4397 (2022).
Henshall, D. C. & Kobow, K. Epigenetics and epilepsy. Cold Spring Harb Perspect Med. 5, https://doi.org/10.1101/cshperspect.a022731 (2015).
Dhureja, M., Chaturvedi, P., Choudhary, A., Kumar, P. & Munshi, A. Molecular insights of drug resistance in epilepsy: multi-omics unveil. Mol. Neurobiol. 62, https://doi.org/10.1007/s12035-024-04220-6 (2025).
Luo, Y.-F. et al. Divergent projections of the prelimbic cortex mediate autism- and anxiety-like behaviors. Mol. Psychiatry 28, 2343–2354 (2023).
Sun, Y. et al. TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis. 8, e3031 (2017).
Mochel, F. et al. Variants in the SK2 channel gene (KCNN2) lead to dominant neurodevelopmental movement disorders. Brain 143, 3564–3573 (2020).
Cho, L. T. Y. et al. An intracellular allosteric modulator binding pocket in SK2 ion channels is shared by multiple chemotypes. Structure 26 https://doi.org/10.1016/j.str.2018.02.017 (2018).
Pascual Cuadrado, D., Wierczeiko, A., Hewel, C., Gerber, S. & Lutz, B. Dichotomic hippocampal transcriptome after glutamatergic vs. GABAergic deletion of the cannabinoid CB1 receptor. Front. Synaptic Neurosci. 13, 660718 (2021).
Gokce-Samar, Z. et al. Molecular and phenotypic characterization of the rorb-related disorder. Neurology 102, e207945 (2024).
Vuong, C. K. et al. Rbfox1 regulates synaptic transmission through the inhibitory neuron-specific vSNARE Vamp1. Neuron 98, https://doi.org/10.1016/j.neuron.2018.03.008 (2018).
Ma, M.-G. et al. RYR2 mutations are associated with benign epilepsy of childhood with centrotemporal spikes with or without arrhythmia. Front. Neurosci. 15, 629610 (2021).
Wang, S. et al. A novel BCL11A polymorphism influences gene expression, therapeutic response and epilepsy risk: a multicenter study. Front. Mol. Neurosci. 15, 1010101 (2022).
Hein, R. F. C. et al. R-SPONDIN2+ mesenchymal cells form the bud tip progenitor niche during human lung development. Dev. Cell 57, https://doi.org/10.1016/j.devcel.2022.05.010 (2022).
Chen, Y., Xu, C., Harirforoosh, S., Luo, X. & Wang, K.-S. Analysis of PTPRK polymorphisms in association with risk and age at onset of Alzheimer’s disease, cancer risk, and cholesterol. J. Psychiatr. Res. 96, 65–72 (2018).
Johannesen, K. M. et al. Genotype-phenotype correlations in SCN8A-related disorders reveal prognostic and therapeutic implications. Brain 145, 2991–3009 (2022).
Yang, N. et al. Antioxidants targeting mitochondrial oxidative stress: promising neuroprotectants for epilepsy. Oxid. Med. Cell Longev. 2020, 6687185 (2020).
Liu, H. et al. Prohibitin 1 regulates mtDNA release and downstream inflammatory responses. EMBO J. 41, e111173 (2022).
Fulton, R. E. et al. Neuron-specific mitochondrial oxidative stress results in epilepsy, glucose dysregulation and a striking astrocyte response. Neurobiol. Dis. 158, 105470 (2021).
Skwarzynska, D., Sun, H., Williamson, J., Kasprzak, I. & Kapur, J. Glycolysis regulates neuronal excitability via lactate receptor, HCA1R. Brain 146, 1888–1902 (2023).
Kumar, A. et al. NaCT/SLC13A5 facilitates citrate import and metabolism under nutrient-limited conditions. Cell Rep. 36, 109701 (2021).
Qiao, Y.-N. et al. Ketogenic diet-produced β-hydroxybutyric acid accumulates brain GABA and increases GABA/glutamate ratio to inhibit epilepsy. Cell Discov. 10, 17 (2024).
Rho, J. M. & Boison, D. The metabolic basis of epilepsy. Nat. Rev. Neurol. 18, 333–347 (2022).
Cai, J. et al. Assessing the causal association between human blood metabolites and the risk of epilepsy. J. Transl. Med. 20, 437 (2022).
Pugacheva, E. M. et al. BORIS/CTCFL epigenetically reprograms clustered CTCF binding sites into alternative transcriptional start sites. Genome Biol. 25, 40 (2024).
Păun, O. et al. Pioneer factor ASCL1 cooperates with the mSWI/SNF complex at distal regulatory elements to regulate human neural differentiation. Genes Dev. 37, 218–242 (2023).
Li, L., Miao, W., Huang, M., Williams, P. & Wang, Y. Integrated genomic and proteomic analyses reveal novel mechanisms of the methyltransferase SETD2 in renal cell carcinoma development. Mol. Cell Proteom. 18, 437–447 (2019).
Du, X., Wang, Y., Wang, X., Tian, X. & Jing, W. Neural circuit mechanisms of epilepsy: maintenance of homeostasis at the cellular, synaptic, and neurotransmitter levels. Neural Regen. Res. 21, 455–465 (2026).
Xiong, H., Tang, F., Guo, Y., Xu, R. & Lei, P. Neural circuit changes in neurological disorders: evidence from in vivo two-photon imaging. Ageing Res. Rev. 87, 101933 (2023).
Wu, Z. et al. FAM69C functions as a kinase for eIF2α and promotes stress granule assembly. EMBO Rep. 24, e55641 (2023).
Kelvington, B. A. & Abel, T. hnRNPH2 gain-of-function mutations reveal therapeutic strategies and a role for RNA granules in neurodevelopmental disorders. J. Clin. Investig. 133, https://doi.org/10.1172/JCI171499 (2023).
Kiebler, M. A. & Bauer, K. E. RNA granules in flux: dynamics to balance physiology and pathology. Nat. Rev. Neurosci. 25, 711–725 (2024).
Zaidi, D. et al. Forebrain Eml1 depletion reveals early centrosomal dysfunction causing subcortical heterotopia. J. Cell Biol. 223, https://doi.org/10.1083/jcb.202310157 (2024).
Bojja, S. L. et al. Metformin ameliorates the status epilepticus-induced hippocampal pathology through possible mTOR modulation. Inflammopharmacology 29, 137–151 (2021).
Wang, X., Hu, Y. & Xu, R. The pathogenic mechanism of TAR DNA-binding protein 43 (TDP-43) in amyotrophic lateral sclerosis. Neural Regen. Res. 19, 800–806 (2024).
Sharma, D., Wangoo, N. & Sharma, R. K. Ultrasensitive NIR fluorometric assay for inorganic pyrophosphatase detection via Cu2+-PPi interaction using bimetallic Au-Ag nanoclusters. Anal. Chim. Acta 1305, 342584 (2024).
Tong, X. et al. TRPM7 contributes to pyroptosis and its involvement in status epilepticus. J. Neuroinflam. 21, 315 (2024).
Kong, Z., Jiang, J., Deng, M., Deng, M. & Wu, H. Improving epilepsy management by targeting P2 × 7 receptor with ROS/electric responsive nanomicelles. J. Nanobiotechnol. 23, 332 (2025).
Schwer, B. et al. Tandem inactivation of inositol pyrophosphatases Asp1, Siw14, and Aps1 illuminates functional redundancies in inositol pyrophosphate catabolism in fission yeast. mBio 16, e0038925 (2025).
Lankinen, K. et al. Cortical depth profiles of auditory and visual 7 T functional MRI responses in human superior temporal areas. Hum. Brain Mapp. 44, 362–372 (2023).
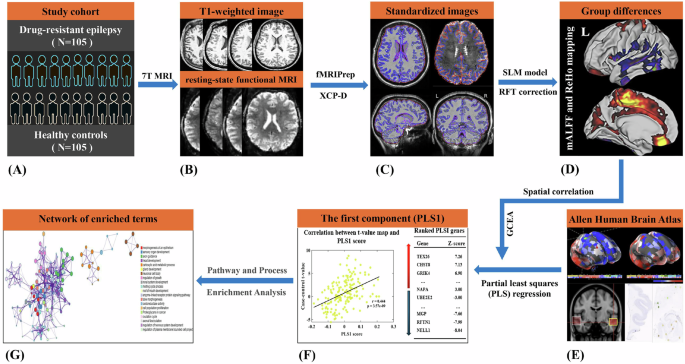
Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019).
Mehta, K. et al. XCP – D: A robust pipeline for the post – processing of fMRI data. Imaging Neurosci (Camb). 2, imag – 2 – 00257 (2024).
Ciric, R. et al. Mitigating head motion artifact in functional connectivity MRI. Nat. Protoc. 13, 2801–2826 (2018).
Worsley, K. J., Taylor, J. E., Tomaiuolo, F. & Lerch, J. Unified univariate and multivariate random field theory. Neuroimage 23, S189–S195 (2004).
Markello, R. D. et al. Standardizing workflows in imaging transcriptomics with the abagen toolbox. Elife 10, https://doi.org/10.7554/eLife.72129 (2021).
Gordon, E. M. et al. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016).
Colombani, C. et al. A comparison of partial least squares (PLS) and sparse PLS regressions in genomic selection in French dairy cattle. J. Dairy Sci. 95, 2120–2131 (2012).
Romero-Garcia, R. et al. Schizotypy-related magnetization of cortex in healthy adolescence is colocated with expression of schizophrenia-related genes. Biol. Psychiatry 88, 248–259 (2020).
Morgan, S. E. et al. Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc. Natl. Acad. Sci. USA 116, 9604–9609 (2019).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Mao, H. Decoding of regional cortical vulnerability to drug-resistant epilepsy using 7T MRI. OSF https://doi.org/10.17605/OSF.IO/6SGWV (2025).