Zhang XX, Tian Y, Wang ZT, Ma YH, Tan L, Yu JT. The epidemiology of alzheimer’s disease modifiable risk factors and prevention. J Prev Alzheimers Dis. 2021;8:313–21.
Farfel JM, Yu L, Boyle PA, Leurgans S, Shah RC, Schneider JA, Bennett DA. Alzheimer’s disease frequency peaks in the tenth decade and is lower afterwards. Acta Neuropathol Commun. 2019;7:104.
Huang H-Z, Wang X, Liu D. Cognition damage due to disruption of cyclic adenosine monophosphate-related signaling pathway in melatonin receptor 2 knockout mice. AN 2023, 2.
Hampel H, Hardy J, Blennow K, Chen C, Perry G, Kim SH, Villemagne VL, Aisen P, Vendruscolo M, Iwatsubo T, et al. The Amyloid-beta pathway in alzheimer’s disease. Mol Psychiatry. 2021;26:5481–503.
Liu D-Y, Huang H-Z, Li K, Lu Y, Zhu L-Q. EPAC2 knockout causes abnormal Tau pathology through calpain-mediated CDK5 activation. AN 2022, 1.
Arnsten AFT, Datta D, Del Tredici K, Braak H. Hypothesis: Tau pathology is an initiating factor in sporadic alzheimer’s disease. Alzheimers Dement. 2021;17:115–24.
Leng F, Edison P. Neuroinflammation and microglial activation in alzheimer disease: where do we go from here? Nat Rev Neurol. 2021;17:157–72.
Rawat P, Sehar U, Bisht J, Selman A, Culberson J, Reddy PH. Phosphorylated Tau in Alzheimer’s Disease and Other Tauopathies. Int J Mol Sci 2022, 23.
Lecca D, Jung YJ, Scerba MT, Hwang I, Kim YK, Kim S, Modrow S, Tweedie D, Hsueh SC, Liu D, et al. Role of chronic neuroinflammation in neuroplasticity and cognitive function: A hypothesis. Alzheimers Dement. 2022;18:2327–40.
Mucke L, Selkoe DJ. Neurotoxicity of amyloid beta-protein: synaptic and network dysfunction. Cold Spring Harb Perspect Med. 2012;2:a006338.
Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with Familial alzheimer’s disease. Nature. 1991;349:704–6.
Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset Familial alzheimer’s disease. Nature. 1995;375:754–60.
Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. Candidate gene for the chromosome 1 Familial alzheimer’s disease locus. Science. 1995;269:973–7.
Kamboh MI. A brief synopsis on the genetics of alzheimer’s disease. Curr Genet Med Rep. 2018;6:133–5.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with alzheimer’s disease. Nat Genet. 2009;41:1088–93.
Andrews SJ, Fulton-Howard B, Goate A. Interpretation of risk loci from genome-wide association studies of alzheimer’s disease. Lancet Neurol. 2020;19:326–35.
Andrieu S, Coley N, Lovestone S, Aisen PS, Vellas B. Prevention of sporadic alzheimer’s disease: lessons learned from clinical trials and future directions. Lancet Neurol. 2015;14:926–44.
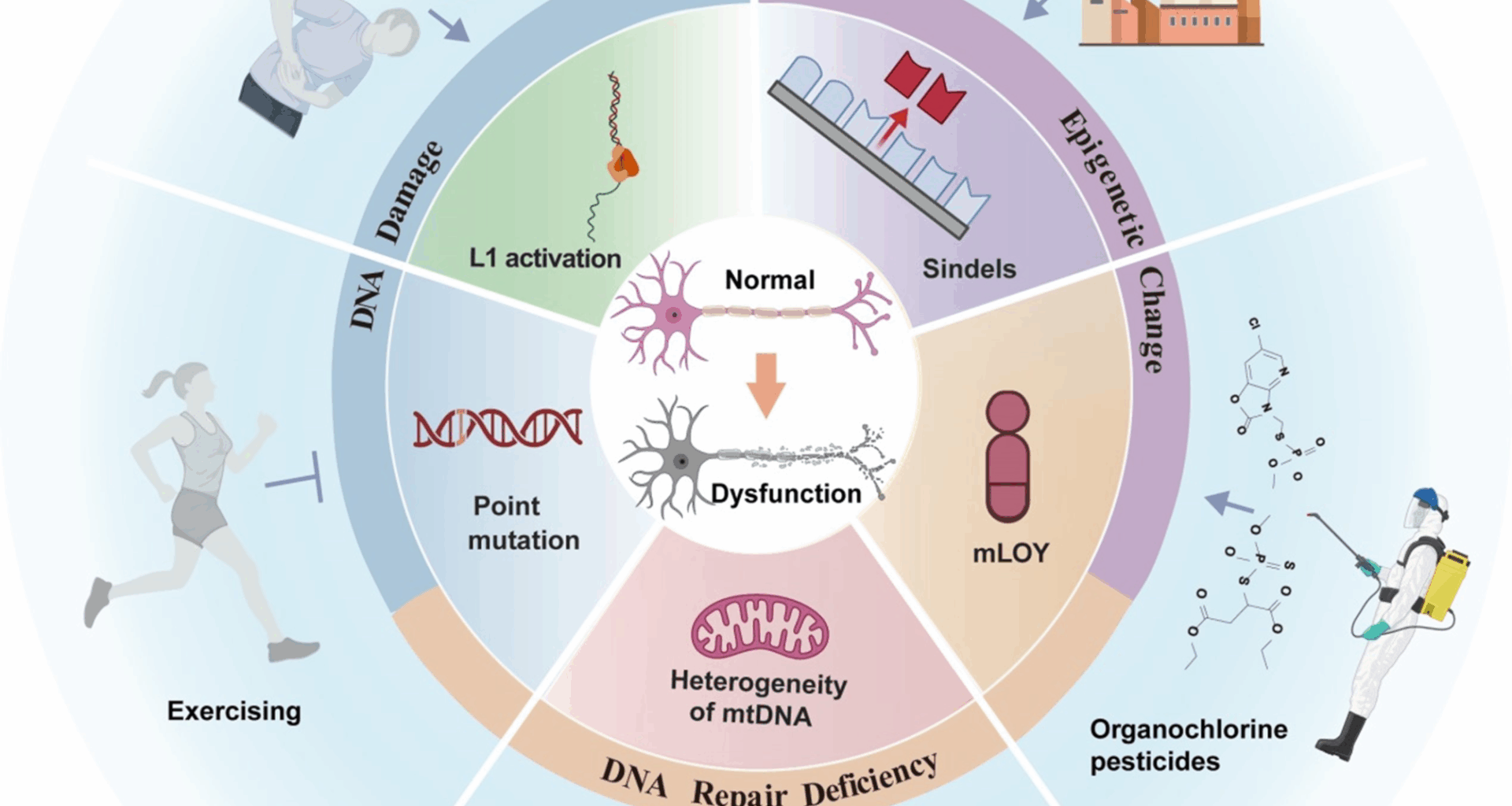
Dhapola R, Sharma P, Kumari S, Bhatti JS, HariKrishnaReddy D. Environmental toxins and alzheimer’s disease: a comprehensive analysis of pathogenic mechanisms and therapeutic modulation. Mol Neurobiol. 2024;61:3657–77.
Li R, Di L, Li J, Fan W, Liu Y, Guo W, Liu W, Liu L, Li Q, Chen L, et al. A body map of somatic mutagenesis in morphologically normal human tissues. Nature. 2021;597:398–403.
Pamphlett R. Somatic mutation: a cause of sporadic neurodegenerative diseases? Med Hypotheses. 2004;62:679–82.
Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, HA FS, Doebley AL, Bernier RA, Nickerson DA. Eichler EE: denovo-db: a compendium of human de Novo variants. Nucleic Acids Res. 2017;45:D804–11.
Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–84.
De S. Somatic mosaicism in healthy human tissues. Trends Genet. 2011;27:217–23.
Vijg J, Dong X. Pathogenic mechanisms of somatic mutation and genome mosaicism in aging. Cell. 2020;182:12–23.
Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O, Bonet J, Kranas H, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–72.
Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–95.
Mass E, Jacome-Galarza CE, Blank T, Lazarov T, Durham BH, Ozkaya N, Pastore A, Schwabenland M, Chung YR, Rosenblum MK, et al. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 2017;549:389–93.
Aldous SG, Smith EJ, Landles C, Osborne GF, Canibano-Pico M, Nita IM, Phillips J, Zhang Y, Jin B, Hirst MB, et al. A CAG repeat threshold for therapeutics targeting somatic instability in huntington’s disease. Brain. 2024;147:1784–98.
Handsaker RE, Kashin S, Reed NM, Tan S, Lee WS, McDonald TM, Morris K, Kamitaki N, Mullally CD, Morakabati NR, et al. Long somatic DNA-repeat expansion drives neurodegeneration in huntington’s disease. Cell. 2025;188:623–e639619.
Chahal G, Tyagi S, Ramialison M. Navigating the non-coding genome in heart development and congenital heart disease. Differentiation. 2019;107:11–23.
Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME. McVean GA: A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77.
Ganz J, Luquette LJ, Bizzotto S, Miller MB, Zhou Z, Bohrson CL, Jin H, Tran AV, Viswanadham VV, McDonough G, et al. Contrasting somatic mutation patterns in aging human neurons and oligodendrocytes. Cell. 2024;187:1955–70. e1923.
Huang AY, Zhou Z, Talukdar M, Miller MB, Chhouk B, Enyenihi L, Rosen I, Stronge E, Zhao B, Kim D et al. Somatic cancer driver mutations are enriched and associated with inflammatory states in Alzheimer’s disease microglia. bioRxiv 2024.
Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen BJ, Venturini E, et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018;359:550–5.
Luquette LJ, Miller MB, Zhou Z, Bohrson CL, Zhao Y, Jin H, Gulhan D, Ganz J, Bizzotto S, Kirkham S, et al. Single-cell genome sequencing of human neurons identifies somatic point mutation and indel enrichment in regulatory elements. Nat Genet. 2022;54:1564–71.
Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33.
Miller MB, Huang AY, Kim J, Zhou Z, Kirkham SL, Maury EA, Ziegenfuss JS, Reed HC, Neil JE, Rento L, et al. Somatic genomic changes in single alzheimer’s disease neurons. Nature. 2022;604:714–22.
Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–6.
Hazen JL, Faust GG, Rodriguez AR, Ferguson WC, Shumilina S, Clark RA, Boland MJ, Martin G, Chubukov P, Tsunemoto RK, et al. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron. 2016;89:1223–36.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87.
Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–8.
Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 Retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–96.
Hoang ML, Kinde I, Tomasetti C, McMahon KW, Rosenquist TA, Grollman AP, Kinzler KW, Vogelstein B, Papadopoulos N. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc Natl Acad Sci U S A. 2016;113:9846–51.
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85.
Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Luquette LJ, Yandava CN, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359:555–9.
Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–7.
Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, Gomez C, Degasperi A, Harris R, Jackson SP, et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177:821–e836816.
Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-Electron reduction potentials of adenosine and Guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–8.
Osorio FG, Rosendahl Huber A, Oka R, Verheul M, Patel SH, Hasaart K, de la Fonteijne L, Varela I, Camargo FD, van Boxtel R. Somatic mutations reveal lineage relationships and Age-Related mutagenesis in human hematopoiesis. Cell Rep. 2018;25:2308–e23162304.
Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–950.
Zhou Z, Luquette LJ, Dong G, Kim J, Ku J, Kim K, Bae M, Shao DD, Sahile B, Miller MB et al. Recurrent patterns of widespread neuronal genomic damage shared by major neurodegenerative disorders. bioRxiv 2025.
Reijns MAM, Parry DA, Williams TC, Nadeu F, Hindshaw RL, Rios Szwed DO, Nicholson MD, Carroll P, Boyle S, Royo R, et al. Signatures of TOP1 transcription-associated mutagenesis in cancer and germline. Nature. 2022;602:623–31.
Jin B, Brown KS, Smirnov DS, Naik SM, Kirkham SL, Hennessey EL, Dong G, Mao S, Wijethunga SP, Stewart TC et al. Neurons accumulate disease-specific somatic genomic changes across tau pathologic states in Alzheimer’s disease. bioRxiv 2025.
Dong G, Ma CC, Mao S, Naik SM, Brown KS, McDonough GA, Kim J, Kirkham SL, Cherry JD, Uretsky M et al. Diverse somatic genomic alterations in single neurons in chronic traumatic encephalopathy. bioRxiv 2025.
Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101.
Consortium ITP-CAWG. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93.
Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65.
Chronister WD, Burbulis IE, Wierman MB, Wolpert MJ, Haakenson MF, Smith ACB, Kleinman JE, Hyde TM, Weinberger DR, Bekiranov S, McConnell MJ. Neurons with complex karyotypes are rare in aged human neocortex. Cell Rep. 2019;26:825–e835827.
Sun C, Kathuria K, Emery SB, Kim B, Burbulis IE, Shin JH, Brain Somatic Mosaicism N, Weinberger DR, Moran JV, Kidd JM, et al. Mapping recurrent mosaic copy number variation in human neurons. Nat Commun. 2024;15:4220.
Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000.
Kim J, Huang AY, Johnson SL, Lai J, Isacco L, Jeffries AM, Miller MB, Lodato MA, Walsh CA, Lee EA. Prevalence and mechanisms of somatic deletions in single human neurons during normal aging and in DNA repair disorders. Nat Commun. 2022;13:5918.
Knouse KA, Wu J, Amon A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 2016;26:376–84.
McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH. Mosaic copy number variation in human neurons. Science. 2013;342:632–7.
Sekar S, Tomasini L, Proukakis C, Bae T, Manlove L, Jang Y, Scuderi S, Zhou B, Kalyva M, Amiri A, et al. Complex mosaic structural variations in human fetal brains. Genome Res. 2020;30:1695–704.
Piazza A, Heyer WD. Homologous recombination and the formation of complex genomic rearrangements. Trends Cell Biol. 2019;29:135–49.
Savocco J, Piazza A. Recombination-mediated genome rearrangements. Curr Opin Genet Dev. 2021;71:63–71.
Pascarella G, Hon CC, Hashimoto K, Busch A, Luginbuhl J, Parr C, Hin Yip W, Abe K, Kratz A, Bonetti A, et al. Recombination of repeat elements generates somatic complexity in human genomes. Cell. 2022;185:3025–e30403026.
Malumbres M, Villarroya-Beltri C. Mosaic variegated aneuploidy in development, ageing and cancer. Nat Rev Genet. 2024;25:864–78.
Pack SD, Weil RJ, Vortmeyer AO, Zeng W, Li J, Okamoto H, Furuta M, Pak E, Lubensky IA, Oldfield EH, Zhuang Z. Individual adult human neurons display aneuploidy: detection by fluorescence in situ hybridization and single neuron PCR. Cell Cycle. 2005;4:1758–60.
Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–80.
Yurov YB, Iourov IY, Monakhov VV, Soloviev IV, Vostrikov VM, Vorsanova SG. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem. 2005;53:385–90.
van den Bos H, Spierings DC, Taudt AS, Bakker B, Porubsky D, Falconer E, Novoa C, Halsema N, Kazemier HG, Hoekstra-Wakker K, et al. Single-cell whole genome sequencing reveals no evidence for common aneuploidy in normal and alzheimer’s disease neurons. Genome Biol. 2016;17:116.
Abou Abdallah F, Abdel Massih C, Attieh C, Chebly A. The impact of mosaic loss of the Y chromosome (mLOY) in men of advanced age. Biogerontology. 2024;25:943–55.
Vermeulen MC, Pearse R, Young-Pearse T, Mostafavi S. Mosaic loss of chromosome Y in aged human microglia. Genome Res. 2022;32:1795–807.
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82.
Xie Y, Mates L, Ivics Z, Izsvak Z, Martin SL, An W. Cell division promotes efficient Retrotransposition in a stable L1 reporter cell line. Mob DNA. 2013;4:10.
Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH. Jr.: L1 Retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12.
Macia A, Widmann TJ, Heras SR, Ayllon V, Sanchez L, Benkaddour-Boumzaouad M, Munoz-Lopez M, Rubio A, Amador-Cubero S, Blanco-Jimenez E, et al. Engineered LINE-1 Retrotransposition in nondividing human neurons. Genome Res. 2017;27:335–48.
Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. 2014;48:1–27.
Peze-Heidsieck E, Bonnifet T, Znaidi R, Ravel-Godreuil C, Massiani-Beaudoin O, Joshi RL, Fuchs J. Retrotransposons as a source of DNA damage in neurodegeneration. Front Aging Neurosci. 2021;13:786897.
Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–39.
Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, Walsh CA. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59.
Sanchez-Luque FJ, Kempen MHC, Gerdes P, Vargas-Landin DB, Richardson SR, Troskie RL, Jesuadian JS, Cheetham SW, Carreira PE, Salvador-Palomeque C, et al. LINE-1 evasion of epigenetic repression in humans. Mol Cell. 2019;75:590–e604512.
Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–23.
Prigione A, Lichtner B, Kuhl H, Struys EA, Wamelink M, Lehrach H, Ralser M, Timmermann B, Adjaye J. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell-like metabolic reprogramming. Stem Cells. 2011;29:1338–48.
Pursell ZF, McDonald JT, Mathews CK, Kunkel TA. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity. Nucleic Acids Res. 2008;36:2174–81.
Marcelino LA, Thilly WG. Mitochondrial mutagenesis in human cells and tissues. Mutat Res. 1999;434:177–203.
Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–13.
Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24.
Smith ALM, Whitehall JC, Greaves LC. Mitochondrial DNA mutations in ageing and cancer. Mol Oncol. 2022;16:3276–94.
Li H, Slone J, Fei L, Huang T. Mitochondrial DNA Variants and Common Diseases: A Mathematical Model for the Diversity of Age-Related mtDNA Mutations. Cells. 2019, 8.
Klein HU, Trumpff C, Yang HS, Lee AJ, Picard M, Bennett DA, De Jager PL. Characterization of mitochondrial DNA quantity and quality in the human aged and alzheimer’s disease brain. Mol Neurodegener. 2021;16:75.
Hoekstra JG, Hipp MJ, Montine TJ, Kennedy SR. Mitochondrial DNA mutations increase in early stage alzheimer disease and are inconsistent with oxidative damage. Ann Neurol. 2016;80:301–6.
Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–1.
Zheng W, Khrapko K, Coller HA, Thilly WG, Copeland WC. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat Res. 2006;599:11–20.
Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706.
Anderson AP, Luo X, Russell W, Yin YW. Oxidative damage diminishes mitochondrial DNA polymerase replication fidelity. Nucleic Acids Res. 2020;48:817–29.
Hu H, Li M. Mitochondria-targeted antioxidant Mitotempo protects mitochondrial function against amyloid beta toxicity in primary cultured mouse neurons. Biochem Biophys Res Commun. 2016;478:174–80.
Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794.
Yuan Y, Ju YS, Kim Y, Li J, Wang Y, Yoon CJ, Yang Y, Martincorena I, Creighton CJ, Weinstein JN, et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52:342–52.
Graziewicz MA, Bienstock RJ, Copeland WC. The DNA polymerase gamma Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2’-deoxyguanosine. Hum Mol Genet. 2007;16:2729–39.
Xu Y, Xu L, Han M, Liu X, Li F, Zhou X, Wang Y, Bi J. Altered mitochondrial DNA methylation and mitochondrial DNA copy number in an APP/PS1 Transgenic mouse model of alzheimer disease. Biochem Biophys Res Commun. 2019;520:41–6.
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93.
Moore L, Cagan A, Coorens THH, Neville MDC, Sanghvi R, Sanders MA, Oliver TRW, Leongamornlert D, Ellis P, Noorani A, et al. The mutational landscape of human somatic and germline cells. Nature. 2021;597:381–6.
Wong JKL, Aichmuller C, Schulze M, Hlevnjak M, Elgaafary S, Lichter P, Zapatka M. Association of mutation signature effectuating processes with mutation hotspots in driver genes and non-coding regions. Nat Commun. 2022;13:178.
Li L, Jiang D, Zhang Q, Liu H, Xu F, Guo C, Qin Z, Wang H, Feng J, Liu Y, et al. Integrative proteogenomic characterization of early esophageal cancer. Nat Commun. 2023;14:1666.
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81.
Dileep V, Boix CA, Mathys H, Marco A, Welch GM, Meharena HS, Loon A, Jeloka R, Peng Z, Bennett DA, et al. Neuronal DNA double-strand breaks lead to genome structural variations and 3D genome disruption in neurodegeneration. Cell. 2023;186:4404–e44214420.
Newell ME, Babbrah A, Aravindan A, Rathnam R, Kiernan R, Driver EM, Bowes DA, Halden RU. Prevalence rates of neurodegenerative diseases versus human exposures to heavy metals across the united States. Sci Total Environ. 2024;928:172260.
Yu F, Vock DM, Zhang L, Salisbury D, Nelson NW, Chow LS, Smith G, Barclay TR, Dysken M, Wyman JF. Cognitive effects of aerobic exercise in alzheimer’s disease: A pilot randomized controlled trial. J Alzheimers Dis. 2021;80:233–44.
De la Rosa A, Olaso-Gonzalez G, Arc-Chagnaud C, Millan F, Salvador-Pascual A, Garcia-Lucerga C, Blasco-Lafarga C, Garcia-Dominguez E, Carretero A, Correas AG, et al. Physical exercise in the prevention and treatment of alzheimer’s disease. J Sport Health Sci. 2020;9:394–404.
Luchsinger JA, Mayeux R. Dietary factors and alzheimer’s disease. Lancet Neurol. 2004;3:579–87.
Yan D, Zhang Y, Liu L, Yan H. Pesticide exposure and risk of alzheimer’s disease: a systematic review and meta-analysis. Sci Rep. 2016;6:32222.
Mostafalou S, Abdollahi M. The link of organophosphorus pesticides with neurodegenerative and neurodevelopmental diseases based on evidence and mechanisms. Toxicology. 2018;409:44–52.
Huang D, Chen L, Ji Q, Xiang Y, Zhou Q, Chen K, Zhang X, Zou F, Zhang X, Zhao Z, et al. Lead aggravates alzheimer’s disease pathology via mitochondrial copper accumulation regulated by COX17. Redox Biol. 2024;69:102990.
Li Z, Liang D, Ebelt S, Gearing M, Kobor MS, Konwar C, Maclsaac JL, Dever K, Wingo AP, Levey AI, et al. Differential DNA methylation in the brain as potential mediator of the association between traffic-related PM(2.5) and neuropathology markers of alzheimer’s disease. Alzheimers Dement. 2024;20:2538–51.
Yadav B, Kaur S, Yadav A, Verma H, Kar S, Sahu BK, Pati KR, Sarkar B, Dhiman M, Mantha AK. Implications of organophosphate pesticides on brain cells and their contribution toward progression of alzheimer’s disease. J Biochem Mol Toxicol. 2024;38:e23660.
Chen H, Liu S, Ge B, Zhou D, Li M, Li W, Ma F, Liu Z, Ji Y, Huang G. Effects of folic acid and vitamin B12 supplementation on cognitive impairment and inflammation in patients with alzheimer’s disease: A randomized, Single-Blinded, Placebo-Controlled trial. J Prev Alzheimers Dis. 2021;8:249–56.
Chen H, Liu S, Ji L, Wu T, Ji Y, Zhou Y, Zheng M, Zhang M, Xu W, Huang G. Folic acid supplementation mitigates alzheimer’s disease by reducing inflammation: A randomized controlled trial. Mediators Inflamm. 2016;2016:5912146.
Clark-Matott J, Saleem A, Dai Y, Shurubor Y, Ma X, Safdar A, Beal MF, Tarnopolsky M, Simon DK. Metabolomic analysis of exercise effects in the POLG mitochondrial DNA mutator mouse brain. Neurobiol Aging. 2015;36:2972–83.
Jia D, Tian Z, Wang R. Exercise mitigates age-related metabolic diseases by improving mitochondrial dysfunction. Ageing Res Rev. 2023;91:102087.
Liang J, Wang C, Zhang H, Huang J, Xie J, Chen N. Exercise-Induced benefits for alzheimer’s disease by stimulating mitophagy and improving mitochondrial function. Front Aging Neurosci. 2021;13:755665.
El-Sawy M, Kale SP, Dugan C, Nguyen TQ, Belancio V, Bruch H, Roy-Engel AM, Deininger PL. Nickel stimulates L1 Retrotransposition by a post-transcriptional mechanism. J Mol Biol. 2005;354:246–57.
Balali-Mood M, Naseri K, Tahergorabi Z, Khazdair MR, Sadeghi M. Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front Pharmacol. 2021;12:643972.
Randall JR, Hirst WG, Simmons LA. Substrate specificity for bacterial RNases HII and HIII is influenced by metal availability. J Bacteriol 2018, 200.
Baba M, Kojima K, Nakase R, Imai S, Yamasaki T, Takita T, Crouch RJ, Yasukawa K. Effects of neutral salts and pH on the activity and stability of human RNase H2. J Biochem. 2017;162:211–9.
Sugawara S, Okada R, Loo TM, Tanaka H, Miyata K, Chiba M, Kawasaki H, Katoh K, Kaji S, Maezawa Y, et al. RNaseH2A downregulation drives inflammatory gene expression via genomic DNA fragmentation in senescent and cancer cells. Commun Biol. 2022;5:1420.
Stribinskis V, Ramos KS. Activation of human long interspersed nuclear element 1 Retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 2006;66:2616–20.
Guan X, Meng X, Zhong G, Zhang Z, Wang C, Xiao Y, Fu M, Zhao H, Zhou Y, Hong S, et al. Particulate matter pollution, polygenic risk score and mosaic loss of chromosome Y in middle-aged and older men from the Dongfeng-Tongji cohort study. J Hazard Mater. 2024;471:134315.
Demanelis K, Delgado DA, Tong L, Jasmine F, Ahmed A, Islam T, Parvez F, Kibriya MG, Graziano JH, Ahsan H, Pierce BL. Somatic loss of the Y chromosome is associated with arsenic exposure among Bangladeshi men. Int J Epidemiol. 2023;52:1035–46.
Field MS, Kamynina E, Chon J, Stover PJ. Nuclear folate metabolism. Annu Rev Nutr. 2018;38:219–43.
Walsh DJ, Bernard DJ, Fiddler JL, Pangilinan F, Esposito M, Harold D, Field MS, Parle-McDermott A, Brody LC. Vitamin B12 status and folic acid supplementation influence mitochondrial heteroplasmy levels in mice. PNAS Nexus. 2024;3:pgae116.
Shah HK, Sharma T, Banerjee BD. Organochlorine pesticides induce inflammation, ROS production, and DNA damage in human epithelial ovary cells: an in vitro study. Chemosphere. 2020;246:125691.
Lundqvist J, Hellman B, Oskarsson A. Fungicide Prochloraz induces oxidative stress and DNA damage in vitro. Food Chem Toxicol. 2016;91:36–41.
Tang BL. Neuropathological Mechanisms Associated with Pesticides in Alzheimer’s Disease. Toxics 2020, 8.
Marques-Rocha JL, Milagro FI, Mansego ML, Mourao DM, Martinez JA, Bressan J. LINE-1 methylation is positively associated with healthier lifestyle but inversely related to body fat mass in healthy young individuals. Epigenetics. 2016;11:49–60.
Baylin SB, Ohm JE. Epigenetic gene Silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16.
Soares JP, Silva AM, Oliveira MM, Peixoto F, Gaivao I, Mota MP. Effects of combined physical exercise training on DNA damage and repair capacity: role of oxidative stress changes. Age (Dordr). 2015;37:9799.
Bakulski KM, Seo YA, Hickman RC, Brandt D, Vadari HS, Hu H, Park SK. Heavy metals exposure and alzheimer’s disease and related dementias. J Alzheimers Dis. 2020;76:1215–42.
Lochhead JJ, McCaffrey G, Quigley CE, Finch J, DeMarco KM, Nametz N, Davis TP. Oxidative stress increases blood-brain barrier permeability and induces alterations in occludin during hypoxia-reoxygenation. J Cereb Blood Flow Metab. 2010;30:1625–36.
Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Nucleic acid oxidation in alzheimer disease. Free Radic Biol Med. 2008;44:1493–505.
Gabbita SP, Lovell MA, Markesbery WR. Increased nuclear DNA oxidation in the brain in alzheimer’s disease. J Neurochem. 1998;71:2034–40.
Lovell MA, Soman S, Bradley MA. Oxidatively modified nucleic acids in preclinical alzheimer’s disease (PCAD) brain. Mech Ageing Dev. 2011;132:443–8.
Gwon AR, Park JS, Arumugam TV, Kwon YK, Chan SL, Kim SH, Baik SH, Yang S, Yun YK, Choi Y, et al. Oxidative lipid modification of nicastrin enhances amyloidogenic gamma-secretase activity in alzheimer’s disease. Aging Cell. 2012;11:559–68.
McKinnon PJ. Genome integrity and disease prevention in the nervous system. Genes Dev. 2017;31:1180–94.
Shanbhag NM, Evans MD, Mao W, Nana AL, Seeley WW, Adame A, Rissman RA, Masliah E, Mucke L. Early neuronal accumulation of DNA double strand breaks in alzheimer’s disease. Acta Neuropathol Commun. 2019;7:77.
Thadathil N, Delotterie DF, Xiao J, Hori R, McDonald MP, Khan MM. DNA Double-Strand break accumulation in alzheimer’s disease: evidence from experimental models and postmortem human brains. Mol Neurobiol. 2021;58:118–31.
Zhang X, Liu Y, Huang M, Gunewardena S, Haeri M, Swerdlow RH, Wang N. Landscape of Double-Stranded DNA breaks in postmortem brains from alzheimer’s disease and Non-Demented individuals. J Alzheimers Dis. 2023;94:519–35.
Gorbunova V, Seluanov A. Making ends Meet in old age: DSB repair and aging. Mech Ageing Dev. 2005;126:621–8.
Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–54.
Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–15.
Bader CPJ, Miyazaki-Kasho E, Forslund JME, Dash A, Wessels M, Wanrooij PH. The low endoribonuclease activity and lack of rNMP preference of human mitochondrial topoisomerase 1 protect against ribonucleotide-dependent deletions. Nucleic Acids Res 2025, 53.
Chiu HC, Koh KD, Evich M, Lesiak AL, Germann MW, Bongiorno A, Riedo E, Storici F. RNA intrusions change DNA elastic properties and structure. Nanoscale. 2014;6:10009–17.
DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA Dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–16.
Zhou ZX, Williams JS, Lujan SA, Kunkel TA. Ribonucleotide incorporation into DNA during DNA replication and its consequences. Crit Rev Biochem Mol Biol. 2021;56:109–24.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705.
Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC, et al. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A. 2009;106:6209–14.
Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–7.
Ertuzun T, Semerci A, Cakir ME, Ekmekcioglu A, Gok MO, Soltys DT, de Souza-Pinto NC, Sezerman U, Muftuoglu M. Investigation of base excision repair gene variants in late-onset alzheimer’s disease. PLoS ONE. 2019;14:e0221362.
Mullins EA, Rodriguez AA, Bradley NP, Eichman BF. Emerging roles of DNA glycosylases and the base excision repair pathway. Trends Biochem Sci. 2019;44:765–81.
Kuper J, Kisker C. At the core of nucleotide excision repair. Curr Opin Struct Biol. 2023;80:102605.
Li W, Liu W, Kakoki A, Wang R, Adebali O, Jiang Y, Sancar A. Nucleotide excision repair capacity increases during differentiation of human embryonic carcinoma cells into neurons and muscle cells. J Biol Chem. 2019;294:5914–22.
Sepe S, Payan-Gomez C, Milanese C, Hoeijmakers JH, Mastroberardino PG. Nucleotide excision repair in chronic neurodegenerative diseases. DNA Repair (Amst). 2013;12:568–77.
Zhao B, Rothenberg E, Ramsden DA, Lieber MR. The molecular basis and disease relevance of non-homologous DNA end joining. Nat Rev Mol Cell Biol. 2020;21:765–81.
Pizzamiglio L, Focchi E, Antonucci F. ATM protein kinase: old and new implications in neuronal pathways and brain circuitry. Cells 2020, 9.
Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) Retrotransposition in human neural stem cells. Proc Natl Acad Sci U S A. 2011;108:20382–7.
Bazzani V, Equisoain Redin M, McHale J, Perrone L, Vascotto C. Mitochondrial DNA repair in neurodegenerative diseases and ageing. Int J Mol Sci 2022, 23.
Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–66.
Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–26.
Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–22.
Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–5.
Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaite Z, Muir M, Fluteau A, Aregger M, McEwan A, et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 2018;559:285–9.
Huang SY, Ghosh S, Pommier Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem. 2015;290:14068–76.
Sparks JL, Burgers PM. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 2015;34:1259–69.
Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–81.
Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–4.
Kellner V, Luke B. Molecular and physiological consequences of faulty eukaryotic ribonucleotide excision repair. EMBO J. 2020;39:e102309.
Kazazian HH Jr., Moran JV. Mobile DNA in health and disease. N Engl J Med. 2017;377:361–70.
Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 Retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6.
Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–409.
Ewing AD, Smits N, Sanchez-Luque FJ, Faivre J, Brennan PM, Richardson SR, Cheetham SW, Faulkner GJ. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell. 2020;80:915–e928915.
Guo C, Jeong HH, Hsieh YC, Klein HU, Bennett DA, De Jager PL, Liu Z, Shulman JM. Tau activates transposable elements in alzheimer’s disease. Cell Rep. 2018;23:2874–80.
Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci. 2014;17:357–66.
Sun W, Samimi H, Gamez M, Zare H, Frost B. Pathogenic tau-induced PiRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2018;21:1038–48.
Park SY, Seo J, Chun YS. Targeted downregulation of kdm4a ameliorates Tau-engendered defects in drosophila melanogaster. J Korean Med Sci. 2019;34:e225.
Xiang Y, Yan K, Zheng Q, Ke H, Cheng J, Xiong W, Shi X, Wei L, Zhao M, Yang F, et al. Histone demethylase KDM4B promotes DNA damage by activating long interspersed nuclear Element-1. Cancer Res. 2019;79:86–98.
Kaluski S, Portillo M, Besnard A, Stein D, Einav M, Zhong L, Ueberham U, Arendt T, Mostoslavsky R, Sahay A, Toiber D. Neuroprotective functions for the histone deacetylase SIRT6. Cell Rep. 2017;18:3052–62.
Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011.
Faulkner GJ, Billon V. L1 Retrotransposition in the soma: a field jumping ahead. Mob DNA. 2018;9:22.
Haldar S, Dru C, Mishra R, Tripathi M, Duong F, Angara B, Fernandez A, Arditi M, Bhowmick NA. Histone deacetylase inhibitors mediate DNA damage repair in ameliorating hemorrhagic cystitis. Sci Rep. 2016;6:39257.
Milholland B, Dong X, Zhang L, Hao X, Suh Y, Vijg J. Differences between germline and somatic mutation rates in humans and mice. Nat Commun. 2017;8:15183.
Nesta AV, Tafur D, Beck CR. Hotspots of human mutation. Trends Genet. 2021;37:717–29.
Zylka MJ, Simon JM, Philpot BD. Gene length matters in neurons. Neuron. 2015;86:353–5.
Izydorczyk MB, Kalef-Ezra E, Horner DW, Zheng X, Holmes N, Toffoli M, Sahin ZG, Han Y, Mehta HH, Muzny DM et al. Single cell long read whole genome sequencing reveals somatic transposon activity in human brain. medRxiv 2024.
Soheili-Nezhad S, van der Linden RJ, Olde Rikkert M, Sprooten E, Poelmans G. Long genes are more frequently affected by somatic mutations and show reduced expression in alzheimer’s disease: implications for disease etiology. Alzheimers Dement. 2021;17:489–99.
Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81.
Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133:118–26.
Lalonde S, Stone OA, Lessard S, Lavertu A, Desjardins J, Beaudoin M, Rivas M, Stainier DYR, Lettre G. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS ONE. 2017;12:e0178700.
Yang H, Zhong Y, Peng C, Chen JQ, Tian D. Important role of indels in somatic mutations of human cancer genes. BMC Med Genet. 2010;11:128.
Breuss MW, Yang X, Schlachetzki JCM, Antaki D, Lana AJ, Xu X, Chung C, Chai G, Stanley V, Song Q, et al. Somatic mosaicism reveals clonal distributions of neocortical development. Nature. 2022;604:689–96.
Lodato MA, Walsh CA. Genome aging: somatic mutation in the brain links age-related decline with disease and nominates pathogenic mechanisms. Hum Mol Genet. 2019;28:R197–206.
Avagyan S, Henninger JE, Mannherz WP, Mistry M, Yoon J, Yang S, Weber MC, Moore JL, Zon LI. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science. 2021;374:768–72.
Pamphlett R. The somatic-spread hypothesis for sporadic neurodegenerative diseases. Med Hypotheses. 2011;77:544–7.
McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: the brain somatic mosaicism network. Science 2017, 356.
Paquola ACM, Erwin JA, Gage FH. Insights into the role of somatic mosaicism in the brain. Curr Opin Syst Biol. 2017;1:90–4.
Bizzotto S, Dou Y, Ganz J, Doan RN, Kwon M, Bohrson CL, Kim SN, Bae T, Abyzov A, Network NBSM, et al. Landmarks of human embryonic development inscribed in somatic mutations. Science. 2021;371:1249–53.
Bizzotto S. The human brain through the lens of somatic mosaicism. Front Neurosci. 2023;17:1172469.
Sidiropoulos N, Mardin BR, Rodriguez-Gonzalez FG, Bochkov ID, Garg S, Stutz AM, Korbel JO, Aiden EL, Weischenfeldt J. Somatic structural variant formation is guided by and influences genome architecture. Genome Res. 2022;32:643–55.
Hirsch CD, Springer NM. Transposable element influences on gene expression in plants. Biochim Biophys Acta Gene Regul Mech. 2017;1860:157–65.
Mita P, Boeke JD. How retrotransposons shape genome regulation. Curr Opin Genet Dev. 2016;37:90–100.
Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86.
Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32.
Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506.
Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–14.
Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–44.
Bachiller S, Del-Pozo-Martin Y, Carrion AM. L1 Retrotransposition alters the hippocampal genomic landscape enabling memory formation. Brain Behav Immun. 2017;64:65–70.
Wang Y, Xie Y, Li L, He Y, Zheng D, Yu P, Yu L, Tang L, Wang Y, Wang Z. EZH2 RIP-seq identifies Tissue-specific long Non-coding RNAs. Curr Gene Ther. 2018;18:275–85.
Kokaeva FF, Den’mukhametova SV, Kanapin AA, Godukhin OV, Il’in Y, Ivanov VA. Antisence oligodeoxyribonucleotides for fragments of the reverse transcriptase gene of the LINE-1 element of rats disturb the formation of long-term memory. Dokl Biochem Biophys. 2002;383:93–5.
Lee MH, Siddoway B, Kaeser GE, Segota I, Rivera R, Romanow WJ, Liu CS, Park C, Kennedy G, Long T, Chun J. Somatic APP gene recombination in alzheimer’s disease and normal neurons. Nature. 2018;563:639–45.
Kim J, Zhao B, Huang AY, Miller MB, Lodato MA, Walsh CA, Lee EA. APP gene copy number changes reflect exogenous contamination. Nature. 2020;584:E20–8.
Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44.
van der Bliek AM, Sedensky MM, Morgan PG. Cell biology of the mitochondrion. Genetics. 2017;207:843–71.
Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW. The genetics and pathology of mitochondrial disease. J Pathol. 2017;241:236–50.
Craven L, Alston CL, Taylor RW, Turnbull DM. Recent advances in mitochondrial disease. Annu Rev Genomics Hum Genet. 2017;18:257–75.
Carrozzo R, Tessa A, Vazquez-Memije ME, Piemonte F, Patrono C, Malandrini A, Dionisi-Vici C, Vilarinho L, Villanova M, Schagger H, et al. The T9176G MtDNA mutation severely affects ATP production and results in Leigh syndrome. Neurology. 2001;56:687–90.
A FCL. Mitochondrial metabolism and DNA methylation: a review of the interaction between two genomes. Clin Epigenetics. 2020;12:182.
Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16:530–42.
Picard M, McManus MJ, Gray JD, Nasca C, Moffat C, Kopinski PK, Seifert EL, McEwen BS, Wallace DC. Mitochondrial functions modulate neuroendocrine, metabolic, inflammatory, and transcriptional responses to acute psychological stress. Proc Natl Acad Sci U S A. 2015;112:E6614–6623.
Abascal F, Harvey LMR, Mitchell E, Lawson ARJ, Lensing SV, Ellis P, Russell AJC, Alcantara RE, Baez-Ortega A, Wang Y, et al. Somatic mutation landscapes at single-molecule resolution. Nature. 2021;593:405–10.
Park JS, Lee J, Jung ES, Kim MH, Kim IB, Son H, Kim S, Kim S, Park YM, Mook-Jung I, et al. Brain somatic mutations observed in alzheimer’s disease associated with aging and dysregulation of Tau phosphorylation. Nat Commun. 2019;10:3090.
Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, Revesz T, Davis MB, Wood NW, Collinge J, Tabrizi SJ. Somatic and germline mosaicism in sporadic early-onset alzheimer’s disease. Hum Mol Genet. 2004;13:1219–24.
Min S, Li Z, Shieh A, Giase G, Bao R, Zhang C, Kuney L, Kopp R, Asif H, Alliey-Rodriguez N, et al. Absence of coding somatic single nucleotide variants within well-known candidate genes in late-onset sporadic alzheimer’s disease based on the analysis of multi-omics data. Neurobiol Aging. 2021;108:207–9.
Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R. Inhibition of the ubiquitin-proteasome system in alzheimer’s disease. Proc Natl Acad Sci U S A. 2000;97:9902–6.
Park JH, Shin MJ, Youn GS, Yeo HJ, Yeo EJ, Kwon HJ, Lee LR, Kim NY, Kwon SY, Kim SM et al. PEP-1-PIN1 promotes hippocampal neuronal cell survival by inhibiting cellular ROS and MAPK phosphorylation. Biomedicines 2024, 12.
Dang H, Sui M, He Q, Xie J, Liu Y, Hou P, Ji M. Pin1 inhibitor API-1 sensitizes BRAF-mutant thyroid cancers to BRAF inhibitors by attenuating HER3-mediated feedback activation of MAPK/ERK and PI3K/AKT pathways. Int J Biol Macromol. 2023;248:125867.
Vicario R, Fragkogianni S, Weber L, Lazarov T, Hu Y, Hayashi SY, Craddock B, Socci ND, Alberdi A, Baako A et al. A microglia clonal inflammatory disorder in Alzheimer’s disease. Elife 2025, 13.
Kim JS, Trzebanski S, Shin SH, Schori L, Frumer Friedman GR, Ilani NC, Kadam A, Vicario R, Aust O, Bugaeva P, et al. Clonal hematopoiesis-associated motoric deficits caused by monocyte-derived microglia accumulating in aging mice. Cell Rep. 2025;44:115609.
Prinz M, Jung S, Priller J. Microglia biology: one century of evolving concepts. Cell. 2019;179:292–311.
Yan P, Kim KW, Xiao Q, Ma X, Czerniewski LR, Liu H, Rawnsley DR, Yan Y, Randolph GJ, Epelman S et al. Peripheral monocyte-derived cells counter amyloid plaque pathogenesis in a mouse model of Alzheimer’s disease. J Clin Invest 2022, 132.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98.
Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science 2019, 366.
Shlush LI. Age-related clonal hematopoiesis. Blood. 2018;131:496–504.
Williams N, Lee J, Mitchell E, Moore L, Baxter EJ, Hewinson J, Dawson KJ, Menzies A, Godfrey AL, Green AR, et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature. 2022;602:162–8.
Bouzid H, Belk JA, Jan M, Qi Y, Sarnowski C, Wirth S, Ma L, Chrostek MR, Ahmad H, Nachun D, et al. Clonal hematopoiesis is associated with protection from alzheimer’s disease. Nat Med. 2023;29:1662–70.
Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, Dutkiewicz A, Giefing M, Lewandowska MA, Kozlowski P. Somatic mutations in MiRNA genes in lung Cancer-Potential functional consequences of Non-Coding sequence variants. Cancers (Basel) 2019, 11.
Hornshoj H, Nielsen MM, Sinnott-Armstrong NA, Switnicki MP, Juul M, Madsen T, Sallari R, Kellis M, Orntoft T, Hobolth A, Pedersen JS. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. NPJ Genom Med. 2018;3:1.
Helgadottir HT, Lundin P, Wallen Arzt E, Lindstrom AK, Graff C, Eriksson M. Somatic mutation that affects transcription factor binding upstream of CD55 in the Temporal cortex of a late-onset alzheimer disease patient. Hum Mol Genet. 2019;28:2675–85.
Morgan BP. Complement in the pathogenesis of alzheimer’s disease. Semin Immunopathol. 2018;40:113–24.
Wang Y, Li Y, Dalle Lucca SL, Simovic M, Tsokos GC, Dalle Lucca JJ. Decay accelerating factor (CD55) protects neuronal cells from chemical hypoxia-induced injury. J Neuroinflammation. 2010;7:24.
van Beek J, van Meurs M, t Hart BA, Brok HP, Neal JW, Chatagner A, Harris CL, Omidvar N, Morgan BP, Laman JD, Gasque P. Decay-accelerating factor (CD55) is expressed by neurons in response to chronic but not acute autoimmune central nervous system inflammation associated with complement activation. J Immunol. 2005;174:2353–65.
Li W, Prazak L, Chatterjee N, Gruninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in drosophila. Nat Neurosci. 2013;16:529–31.
Chen H, Zheng X, Xiao D, Zheng Y. Age-associated de-repression of retrotransposons in the drosophila fat body, its potential cause and consequence. Aging Cell. 2016;15:542–52.
Protasova MS, Gusev FE, Grigorenko AP, Kuznetsova IL, Rogaev EI, Andreeva TV. Quantitative analysis of L1-Retrotransposons in alzheimer’s disease and aging. Biochem (Mosc). 2017;82:962–71.
Zhang Q, Zhao L, Yang Y, Li S, Liu Y, Chen C. Mosaic loss of chromosome Y promotes leukemogenesis and clonal hematopoiesis. JCI Insight 2022, 7.
Garcia-Gonzalez P, de Rojas I, Moreno-Grau S, Montrreal L, Puerta R, Alarcon-Martin E, Quintela I, Orellana A, Andrade V, Adami PVM et al. Mendelian Randomisation Confirms the Role of Y-Chromosome Loss in Alzheimer’s Disease Aetiopathogenesis in Men. Int J Mol Sci 2023, 24.
Iourov IY, Vorsanova SG, Liehr T, Yurov YB. Aneuploidy in the normal, alzheimer’s disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol Dis. 2009;34:212–20.
Bushman DM, Kaeser GE, Siddoway B, Westra JW, Rivera RR, Rehen SK, Yung YC, Chun J. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. Elife 2015, 4.
McNaughton D, Knight W, Guerreiro R, Ryan N, Lowe J, Poulter M, Nicholl DJ, Hardy J, Revesz T, Lowe J, et al. Duplication of amyloid precursor protein (APP), but not prion protein (PRNP) gene is a significant cause of early onset dementia in a large UK series. Neurobiol Aging. 2012;33:e426413–421.
Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–83.
Liu YY, Tanikawa C, Ueda K, Matsuda K. INKA2, a novel p53 target that interacts with the serine/threonine kinase PAK4. Int J Oncol. 2019;54:1907–20.
Lai CH, Barik P, Hsieh DJ, Day CH, Ho TJ, Chen RJ, Kuo WW, Padma VV, Shibu MA, Huang CY. Inhibition of cell death-inducing p53 target 1 through miR-210-3p overexpression attenuates reactive oxygen species and apoptosis in rat adipose-derived stem cells challenged with angiotensin II. Biochem Biophys Res Commun. 2020;532:347–54.
Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules 2020, 10.
Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–5.
Yin C, Li DY, Guo X, Cao HY, Chen YB, Zhou F, Ge NJ, Liu Y, Guo SS, Zhao Z, et al. NGS-based profiling reveals a critical contributing role of somatic D-loop MtDNA mutations in HBV-related hepatocarcinogenesis. Ann Oncol. 2019;30:953–62.
Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC. Systemic mitochondrial dysfunction and the etiology of alzheimer’s disease and down syndrome dementia. J Alzheimers Dis. 2010;20(Suppl 2):S293–310.
Carvalho G, Repoles BM, Mendes I, Wanrooij PH. Mitochondrial DNA instability in mammalian cells. Antioxid Redox Signal. 2022;36:885–905.
Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic MtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–31.
Wei W, Keogh MJ, Wilson I, Coxhead J, Ryan S, Rollinson S, Griffin H, Kurzawa-Akanbi M, Santibanez-Koref M, Talbot K, et al. Mitochondrial DNA point mutations and relative copy number in 1363 disease and control human brains. Acta Neuropathol Commun. 2017;5:13.
Brown AM, Sheu RK, Mohs R, Haroutunian V, Blass JP. Correlation of the clinical severity of alzheimer’s disease with an aberration in mitochondrial DNA (mtDNA). J Mol Neurosci. 2001;16:41–8.
Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–4.
Yu T, Slone J, Liu W, Barnes R, Opresko PL, Wark L, Mai S, Horvath S, Huang T. Premature aging is associated with higher levels of 8-oxoguanine and increased DNA damage in the Polg mutator mouse. Aging Cell. 2022;21:e13669.
Kukreja L, Kujoth GC, Prolla TA, Van Leuven F, Vassar R. Increased MtDNA mutations with aging promotes amyloid accumulation and brain atrophy in the app/ld Transgenic mouse model of alzheimer’s disease. Mol Neurodegener. 2014;9:16.
Tanaka D, Nakada K, Takao K, Ogasawara E, Kasahara A, Sato A, Yonekawa H, Miyakawa T, Hayashi J. Normal mitochondrial respiratory function is essential for Spatial remote memory in mice. Mol Brain. 2008;1:21.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6.
Gonzalez-Pena V, Natarajan S, Xia Y, Klein D, Carter R, Pang Y, Shaner B, Annu K, Putnam D, Chen W et al. Accurate genomic variant detection in single cells with primary template-directed amplification. Proc Natl Acad Sci U S A 2021, 118.
Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, Xie XS. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science. 2017;356:189–94.
Evrony GD, Hinch AG, Luo C. Applications of Single-Cell DNA sequencing. Annu Rev Genomics Hum Genet. 2021;22:171–97.
Valecha M, Posada D. Somatic variant calling from single-cell DNA sequencing data. Comput Struct Biotechnol J. 2022;20:2978–85.
Ardui S, Ameur A, Vermeesch JR, Hestand MS. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46:2159–68.
Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39:1348–65.
Rajaby R, Liu DX, Au CH, Cheung YT, Lau AYT, Yang QY, Sung WK. INSurVeyor: improving insertion calling from short read sequencing data. Nat Commun. 2023;14:3243.
Ma C, Shi X, Li X, Zhang YP, Peng MS. Comprehensive evaluation and guidance of structural variation detection tools in chicken whole genome sequence data. BMC Genomics. 2024;25:970.
Cameron DL, Di Stefano L, Papenfuss AT. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun. 2019;10:3240.
Yi Z, Zhang X, Tang W, Yu Y, Wei X, Zhang X, Wei W. Strand-selective base editing of human mitochondrial DNA using mitobes. Nat Biotechnol. 2024;42:498–509.
Lim K. Mitochondrial genome editing: strategies, challenges, and applications. BMB Rep. 2024;57:19–29.
Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–7.