Study setting
We conducted an observational cohort study derived from an ongoing, prospective, multicenter surveillance network REA-REZO of ICU-acquired infections and associated risk factors. This cohort included individual patient data from 206 French adult ICUs.
The detailed protocol for data collection and monitoring are available at https://rearezo.chu-lyon.fr/ (website in French).
Study population
The study population consisted of patients admitted in the participating ICU centers between January 1, 2016, and December 31, 2022, who developed a healthcare-associated infection (HAI), defined as a bloodstream infection, pneumonia or central vascular catheter-related infection occurring at least 48 h after ICU admission.
Each ICU admission was treated as a separate observation. The REA-REZO database is anonymized, with each ICU stay assigned with a unique identifier. As a result, if a patient was readmitted to the ICU during the study period, they would be given a new identifier and considered as a distinct observation.
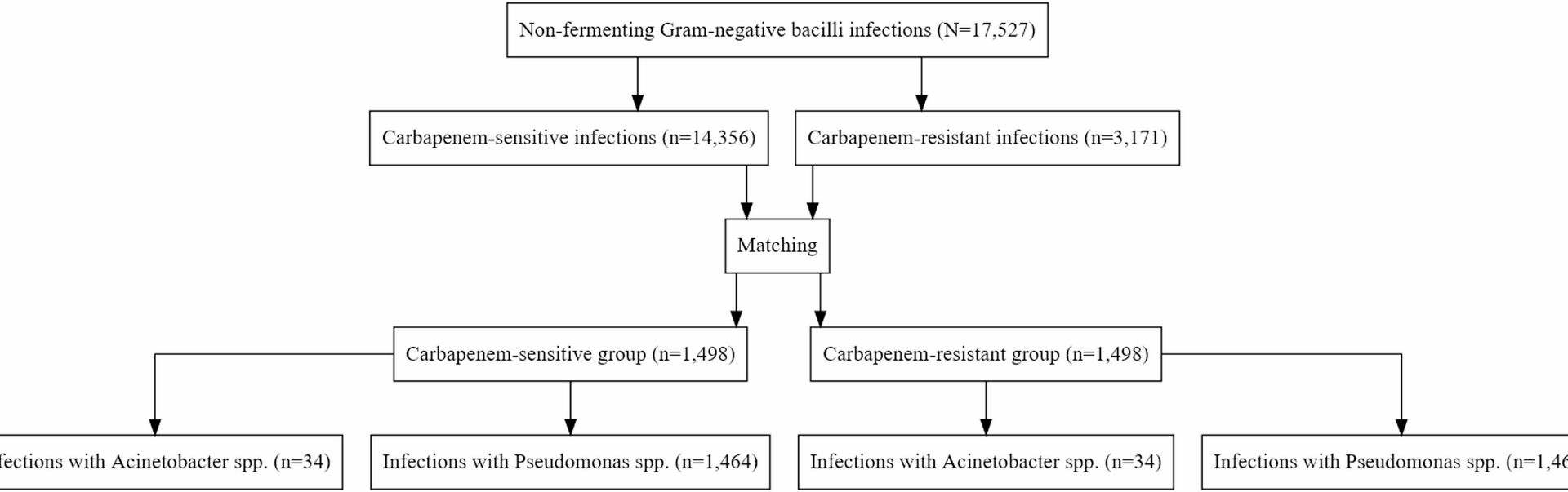
A carbapenem-resistant (CR) Nf-GNB infection was defined as an infection caused by Nf-GNB (Pseudomonas aeruginosa or Acinetobacter baumannii) exhibiting resistance to imipenem and/or meropenem on the antibiotic susceptibility testing results. Antibiotic susceptibility testing, including the detection of carbapenem resistance, was performed according to CA-SFM/EUCAST guidelines [12]. In contrast, carbapenem-susceptible (CS) Nf-GNB was defined as an infection caused by the same pathogens that do not exhibit resistance to these antibiotics.
Data collection
The following data were extracted from the REA-REZO database for analysis: age, sex, Simplified Acute Physiology Score II (SAPS II) measured at admission, ICU admission and discharge dates, vital status at discharge from the ICU, admission category (medical, scheduled or emergency surgery), presence of trauma at admission (defined as any physical injury due to an external cause, regardless of surgical intervention), origin of patient (grouped as community or inter-hospital transfer; nursing home residents were included in the community category), COVID infection at admission, antibiotic treatment within 2 days of admission, mechanical ventilation during the ICU stay, infection date, infectious episode number (defined as the chronological rank of the infection during the ICU stay, i.e., 1st, 2nd, 3rd episode, etc., for each patient), infection site, identified pathogen, resistance to standard antibiotic therapy (ceftazidime and/or piperacillin-tazobactam), and presence of polymicrobial infection. We also collected data on HAI acquired during ventilation defined as a HAI occurring during mechanical ventilation (ventilator-associated pneumonia, bloodstream infection or central vascular catheter-related infection).
Outcomes
Our primary endpoint was 30-day ICU survival. Secondary endpoints included the presence of ICU-acquired reinfection of Nf-GNB, defined as an infection subsequent to another pathogen, and relapse, defined as an infection subsequent to the same pathogen regardless of carbapenem resistance, occurring within 30 days of ICU stay.
Statistical analyses
For each patient with CR Nf-GNB, the first infection was matched at a 1:1 ratio with another ICU patient having CS Nf-GNB infection. Propensity scores were estimated using a logistic regression model, including comprehensive set of covariates known as potential confounders: age, sex, SAPS II, length of ICU stay before infection onset, infectious episode number, infection site, pathogen, year, trauma at admission, admission category, COVID-19 infection at admission, early antibiotic use, patient origin, resistance to piperacillin-tazobactam and/or ceftazidime, polymicrobial infection, and HAI infection during mechanical ventilation [13]. Exact matching was performed on all categorical variables, while matching was performed as closely as possible using a caliper of 0.2 times the standard deviation of the logit of the propensity score for the continuous variables (age, SAPS II and length of ICU stay before infection onset) [14, 15]. The adequacy of matching variables was assessed using the standardized mean difference. The predetermined threshold was set at 0.10 to evaluate the balance achieved in covariate distribution between the CR Nf-GNB and CS Nf-GNB groups. Covariate balance was visually assessed using Love plots (Supplementary Fig. 1) and propensity score distributions before and after matching (Supplementary Fig. 2). Baseline characteristics for the entire unmatched cohort are provided in Supplementary Table 1.
Descriptive statistics were used to report the characteristics of patients with infections caused by CR Nf-GNB compared to those with infections caused by CS Nf-GNB. Quantitative variables were presented as median with interquartile ranges (IQR), while categorical variables were described as counts and proportions (%).
Univariate analyses were performed using the Wilcoxon, Chi-square test or Fisher’s exact test, based on the nature of the variables.
Survival analyses were conducted by taking into account the bias regarding the competing event of ICU discharge, with follow-up starting at the onset of infection, and specific survival at 7, 14, and 30 days were estimated [16]. For reinfection and relapse, the cumulative incidence of the first episode caused by resistant pathogens was analyzed using a competing risks model, with death and ICU discharge as a competing event. Specific cumulative incidences at 7, 14, and 30 days were computed for each group alongside with their 95% confidence interval, and cumulative incidence curves were compared using the Gray test [17].
To assess the association between exposure to CR Nf-GNB infections and the risk of subsequent events in multivariable analyses, competing risks regression models considering discharge a competing event of death were employed [16]. Additionally, for the assessment of relapse, death, discharge, and reinfection were considered as competing risks, while for the assessment of reinfection, death, discharge, and relapse were considered as competing risks.
Sub-Hazard ratios (sHR) were estimated, adjusting for the main confounding factors: age, sex, SAPS II, year of inclusion, admission category, presence of trauma at admission, origin of patient, COVID-19 infection at admission, antibiotic treatment within 2 days of admission, HAI acquired during ventilation, infection site, identified pathogen, length of ICU stay before infection onset, resistance to ceftazidime, resistance to piperacillin-tazobactam, and presence of polymicrobial infection [13]. To evaluate the impact of CR infections on each of the outcomes, a full model incorporating these covariates was initially constructed. A bidirectional stepwise regression based on the Akaike Information Criterion (AIC) was then applied to retain only the relevant confounders in the final model. In order to account for potential dependencies in the data, robust standard errors were computed to estimate the 95% confidence interval. As sensitivity analysis, we also assessed the stability of the estimation on the whole unmatched cohort.
Statistical significance was arbitrarily set at a p-value of 0.05. The analyses were conducted utilizing R software version 4.0.3, incorporating the Survival [18], Matchit [19], and riskRegression packages [20].