Buckley M, Terwagne C, Ganner A, Cubitt L, Brewer R, Kim DK, et al. Saturation genome editing maps the functional spectrum of pathogenic VHL alleles. Nat Genet. 2024;56(7):1446–55.
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–72.
Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–5.
Kierans SJ, Taylor CT. Regulation of glycolysis by the hypoxia-inducible factor (HIF): implications for cellular physiology. J Physiol Internet. 2020. https://doi.org/10.1113/JP280572.
Warburg O, Wind F, Negelein E. The metabolism of tumors in the human body. J Gen Physiol [Internet]. 1927;8(6):519–30.
Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29(5):625–34.
Binderup MLM, Galanakis M, Budtz-Jørgensen E, Kosteljanetz M, Bisgaard ML. Prevalence, birth incidence, and penetrance of von Hippel-Lindau disease (vHL) in Denmark. Eur J Hum Genet. 2017;25(3):301–7.
Maher ER, Iselius L, Yates JRW, Littler M, Benjamin C, Harris R, et al. Von Hippel-Lindau disease: a genetic study. J Med Genet. 1991;28:443–7.
Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. Von Hippel-Lindau disease. Lancet. 2003;361(9374):2059–67.
Neumann HP, Wiestler OD. Clustering of features of von Hippel-Lindau syndrome: evidence for a complex genetic locus. Lancet. 1991;337(8749):1052–4.
Neumann HPH, Eggert HR, Scheremet R, Schumacher M, Mohadjer M, Wakhloo AK, et al. Central nervous system lesions in von Hippel-Lindau syndrome. J Neurol Neurosurg Psychiatry. 1992;55:898–901.
Chen J, Geng W, Zhao Y, Zhao H, Wang G, Huang F, et al. Clinical and mutation analysis of four Chinese families with von Hippel-Lindau disease. Clin Transl Oncol. 2013;15(5):391–7.
Binderup MLM, Jensen AM, Budtz-Jørgensen E, Bisgaard ML. Survival and causes of death in patients with von Hippel-Lindau disease. J Med Genet. 2017;54(1):11–8.
Maher ER, Yates JRW, Harries R, Benjamin C, Harris R, Moore AT, et al. Clinical features and natural history of von hippel-lindau disease. QJM. 1990;77(2):1151–63.
Gordeuk VR, Sergueeva AI, Miasnikova GY, Okhotin D, Voloshin Y, Choyke PL, et al. Congenital disorder of oxygen sensing: association of the homozygous Chuvash polycythemia VHL mutation with thrombosis and vascular abnormalities but not tumors. Blood. 2004;103(10):3924–32.
Perrotta S, Roberti D, Bencivenga D, Corsetto P, O’Brien KA, Caiazza M, et al. Effects of germline VHL deficiency on growth, metabolism, and mitochondria. N Engl J Med. 2020;382(9):835–44.
Flores SK, Cheng Z, Jasper AM, Natori K, Okamoto T, Tanabe A, et al. A synonymous VHL variant in exon 2 confers susceptibility to familial pheochromocytoma and von Hippel-Lindau disease. J Clin Endocrinol Metab. 2019;104(9):3826–34.
Lenglet M, Robriquet F, Schwarz K, Camps C, Couturier A, Hoogewijs D, et al. Identification of a new VHL exon and complex splicing alterations in familial erythrocytosis or von Hippel-Lindau disease. Blood. 2018;132(5):469–83.
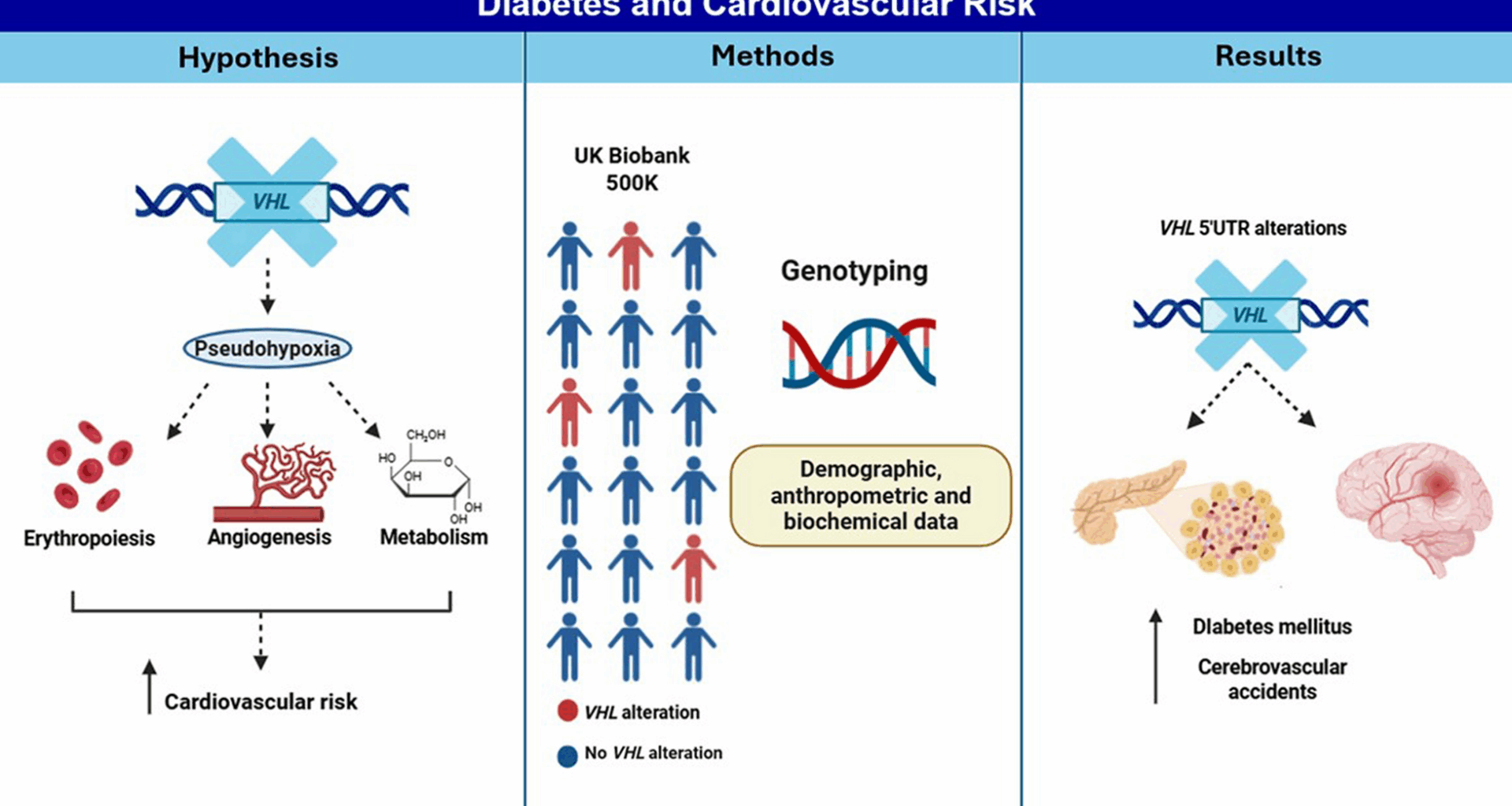
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3): e1001779.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the Association for molecular pathology. Genet Med. 2015;17(5):405–24.
Tirosh A, Sadowski SM, Linehan WM, Libutti SK, Patel D, Nilubol N, et al. Association of VHL genotype with pancreatic neuroendocrine tumor phenotype in patients with von hippel-lindau disease. JAMA Oncol. 2018;4(1):124–6.
Mougel G, Mohamed A, Burnichon N, Giraud S, Pigny P, Bressac-de Paillerets B, et al. Update of the UMD-VHL database: classification of 164 challenging variants based on genotype-phenotype correlation among 605 entries. J Med Genet. 2024;61(4):378–84.
Ou J, Zhu LJ. trackViewer: a Bioconductor package for interactive and integrative visualization of multi-omics data. Nat Methods. 2019;16(6):453–4.
Klimentidis YC, Arora A, Newell M, Zhou J, Ordovas JM, Renquist BJ, et al. Phenotypic and genetic characterization of lower LDL cholesterol and increased type 2 diabetes risk in the UK biobank. Diabetes. 2020;69(10):2194–205.
Iliopoulos O, Levy AP, Jiang C, Kaelin WGJ, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci U S A. 1996;93(20):10595–9.
Zehetner J, Danzer C, Collins S, Eckhardt K, Gerber PA, Ballschmieter P, et al. PVHL is a regulator of glucose metabolism and insulin secretion in pancreatic beta cells. Genes Dev. 2008;22(22):3135–46.
Puri S, Cano DA, Hebrok M. A role for von Hippel-Lindau protein in pancreatic beta-cell function. Diabetes. 2009;58(2):433–41.
Cantley J, Selman C, Shukla D, Abramov AY, Forstreuter F, Esteban MA, et al. Deletion of the von Hippel-Lindau gene in pancreatic beta cells impairs glucose homeostasis in mice. J Clin Invest. 2009;119(1):125–35.
Ilegems E, Bryzgalova G, Correia J, Yesildag B, Berra E, Ruas JL, et al. HIF-1α inhibitor PX-478 preserves pancreatic β cell function in diabetes. Sci Transl Med. 2022;14(638):eaba9112.
Ryczek N, Łyś A, Makałowska I. The functional meaning of 5’UTR in protein-coding genes. Int J Mol Sci. 2023;24(3):2976.
Tamura K, Kanazashi Y, Kawada C, Sekine Y, Maejima K, Ashida S, et al. Variant spectrum of von Hippel-Lindau disease and its genomic heterogeneity in Japan. Hum Mol Genet. 2023;32(12):2046–54.
Chiorean A, Farncombe KM, Delong S, Andric V, Ansar S, Chan C, et al. Large scale genotype- and phenotype-driven machine learning in Von Hippel-Lindau disease. Hum Mutat. 2022;43(9):1268–85.
Mohebian K, Hesse D, Arends D, Brockmann GA. A 5’ UTR mutation contributes to down-regulation of Bbs7 in the Berlin fat mouse. Int J Mol Sci. 2022;23(21):13018.
Ang Z, Paruzzo L, Hayer KE, Schmidt C, Torres Diz M, Xu F, et al. Alternative splicing of its 5’-UTR limits CD20 mRNA translation and enables resistance to CD20-directed immunotherapies. Blood. 2023;142(20):1724–39.
Sergueeva A, Miasnikova G, Shah BN, Song J, Lisina E, Okhotin DJ, et al. Prospective study of thrombosis and thrombospondin-1 expression in Chuvash polycythemia. Vol. 102, Haematologica. Italy; 2017. p. e166–9.
Lorenzo FR, Yang C, Lanikova L, Butros L, Zhuang Z, Prchal JT. Novel compound VHL heterozygosity (VHL T124A/L188V) associated with congenital polycythaemia. Vol. 162, British journal of haematology. England; 2013. p. 851–3.
Chomette L, Migeotte I, Dewachter C, Vachiery JL, Smits G, Bondue A. Early-onset and severe pulmonary arterial hypertension due to a novel compound heterozygous association of rare VHL mutations: A case report and review of existing data. Vol. 12, Pulmonary circulation. United States; 2022. p. e12052.
Gläsker S, Sohn TS, Okamoto H, Li J, Lonser RR, Oldfield EH, et al. Second hit deletion size in von Hippel-Lindau disease. Ann Neurol. 2006;59(1):105–10.
Menendez-Montes I, Escobar B, Palacios B, Gómez MJ, Izquierdo-Garcia JL, Flores L, et al. Myocardial VHL-HIF signaling controls an embryonic metabolic switch essential for cardiac maturation. Dev Cell. 2016;39(6):724–39.
Song SH, Kim JH, Lee JH, Yun YM, Choi DH, Kim HY. Elevated blood viscosity is associated with cerebral small vessel disease in patients with acute ischemic stroke. BMC Neurol. 2017;17(1):20.
Woo HG, Kim HG, Lee KM, Ha SH, Jo H, Heo SH, et al. Blood viscosity associated with stroke mechanism and early neurological deterioration in middle cerebral artery atherosclerosis. Sci Rep. 2023;13(1):9384.
Savatt JM, Kelly MA, Sturm AC, McCormick CZ, Williams MS, Nixon MP, et al. Genomic screening at a single health system. JAMA Netw Open. 2025;8(3): e250917.
Sun Y, Yu Y, Cai L, Yu B, Xiao W, Tan X, et al. Clonal hematopoiesis of indeterminate potential, health indicators, and risk of cardiovascular diseases among patients with diabetes: a prospective cohort study. Cardiovasc Diabetol. 2025;24(1):72.
CDC Summary health statistics [Internet]. Available from: https://www.cdc.gov/nchs/nhis/shs/tables.htm
Ding L, Fan Y, Wang J, Ma X, Chang L, He Q, et al. Central lean mass distribution and the risks of all-cause and cause-specific mortality in 40,283 UK Biobank participants. Obes Facts. 2024;17(5):502–12.
Cheung Q, Wharton S, Josse A, Kuk JL. Ethnic variations in cardiovascular disease (CVD) risk factors and associations with prevalent CVD and CVD mortality in the United States. PLoS ONE. 2025;20(3): e0319617.
Joseph JJ, Ortiz R, Acharya T, Golden SH, López L, Deedwania P. Cardiovascular impact of race and ethnicity in patients with diabetes and obesity: JACC focus seminar 2/9. J Am Coll Cardiol. 2021;78(24):2471–82.
Mital R, Bayne J, Rodriguez F, Ovbiagele B, Bhatt DL, Albert MA. Race and ethnicity considerations in patients with coronary artery disease and stroke: JACC focus seminar 3/9. J Am Coll Cardiol. 2021;78(24):2483–92.