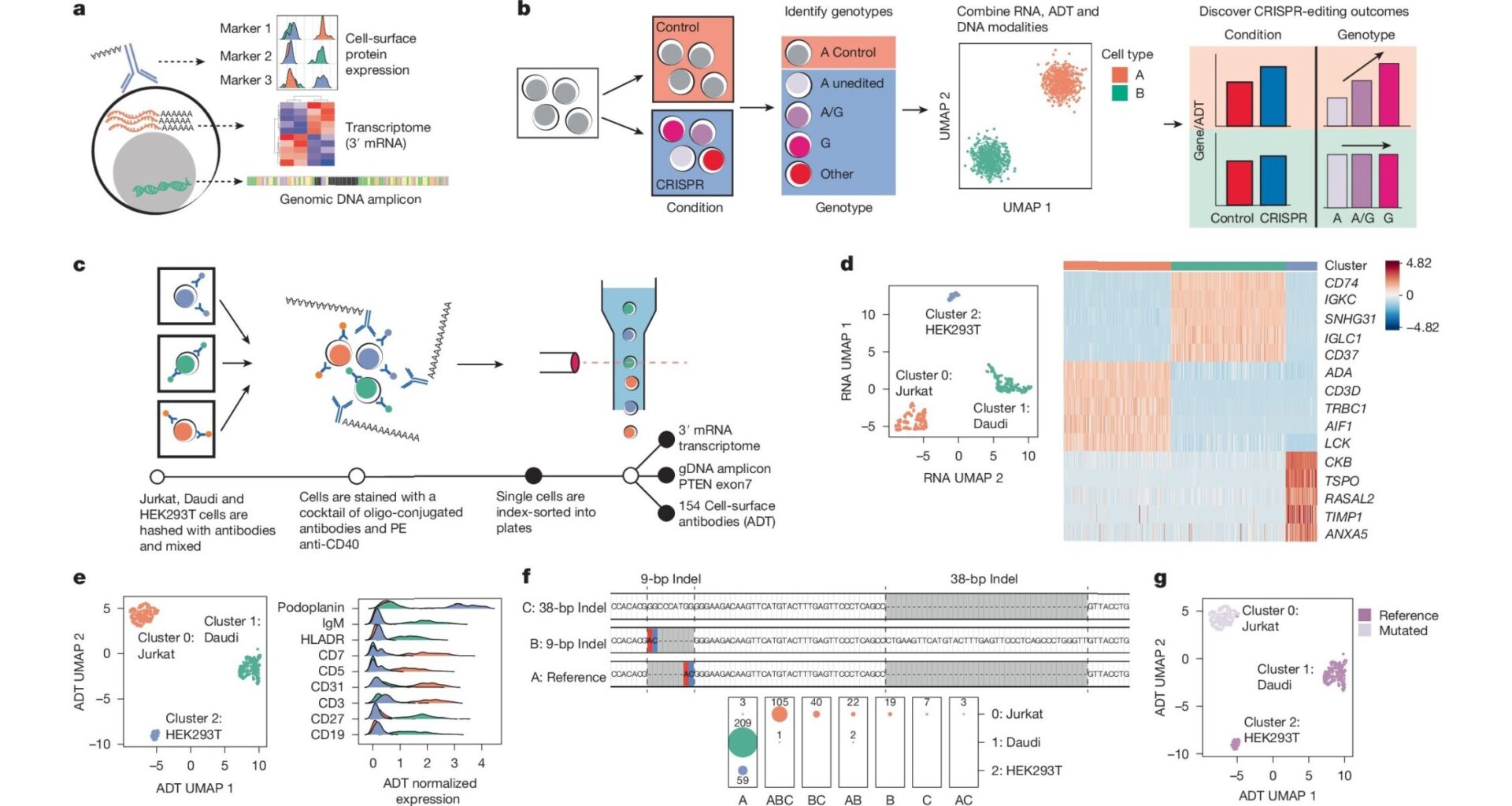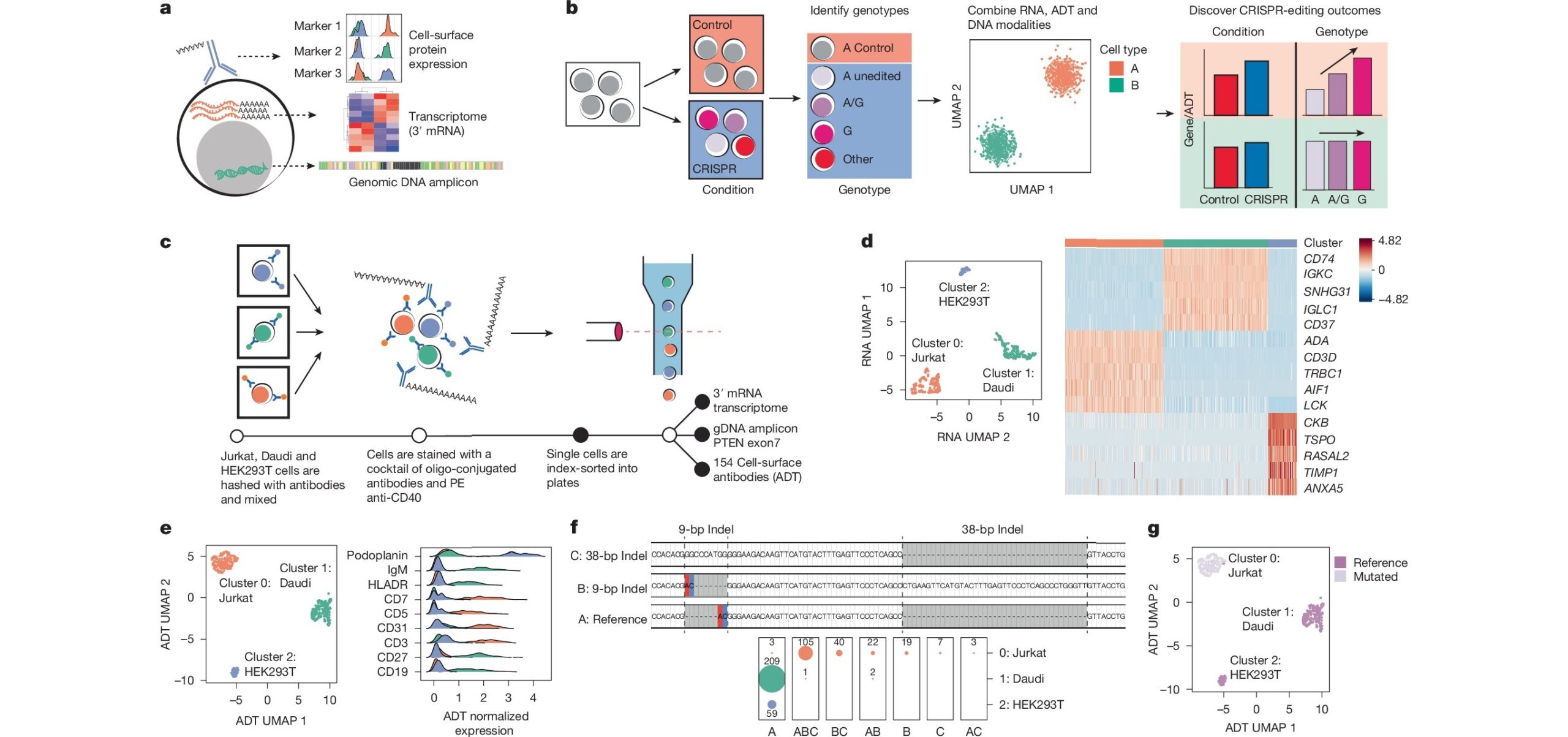
Understanding the genetic drivers of human disease remains a major challenge. Researchers routinely use CRISPR, a gene-editing tool, to model these mutations in complex systems but have not been able to directly examine modified cells.
Now a tool called CRAFTseq, co-developed by an expert now with the Division of Human Genetics at Cincinnati Children’s, will make it easier to understand precisely what happens when disease variants are introduced in model systems.
Details about the new tool were published July 23, 2025 in Nature. Co-corresponding author Yuriy Baglaenko, PhD, conducted the research while with Brigham and Women’s Hospital and the Broad Institute in Boston. Baglaenko has since joined Cincinnati Children’s.
Potential impacts beyond immunology
The tool was used to trace the effects of CRISPR editing in major immune cells. When similar changes were studied using bulk approaches, the research team could not identify statistically significant effects. The results were confounded by samples including a mix of cell populations, secretion of inflammatory cytokines and more.
However, CRAFTseq is a single-cell strategy that accurately models such variables, allowing transcriptional changes and cell surface protein expression within individual cells to be directly attributed to the CRISPR edit. This helps scientists answer questions about whether cell behaviors are truly caused by gene variations or reflect the larger cellular environment.
“We propose that CRAFTseq can be applied on a case-by-case basis to resolve any genetic editing outcome,” the co-authors wrote.
What’s next?
“We are continuing to scale this work with novel microfluidics and to apply the tool directly to primary human B cells to discover genetic determinants of complex autoimmune diseases,” Baglaenko says.
Don’t Miss a Post: