Researchers study how de novo genes may play significant roles in evolution, biology, and disease.
Image credit:Laboratory of Evolutionary Genetics and Genomics at The Rockefeller University
De novo genes are evolution’s magic trick—new genes appearing from non-coding DNA, much like a rabbit pulled from a hat. Once considered impossible, de novo genes have been identified in Drosophila melanogaster, humans, plants, and other organisms.1
One of the researchers leading this work is evolutionary biologist Li Zhao at The Rockefeller University, who has spent much of her career discovering hundreds of de novo genes in fruit flies. Eight years ago, neurophysiologist Torsten Wiesel, Nobel laureate and president emeritus of The Rockefeller University, asked her, “If those new [de novo] genes are genes, how do transcription factors regulate the expression of those genes?” At the time, Zhao didn’t have the tools to answer his question, but she endeavored to find out.
Now in a recent study, published in Nature Ecology & Evolution, Zhao and her team found that about 10 percent of transcription factors (TFs)—including three key TFs—play a major role in regulating de novo gene expression.2 These findings help shed light on the tantalizing mystery of how organisms control new de novo genes.
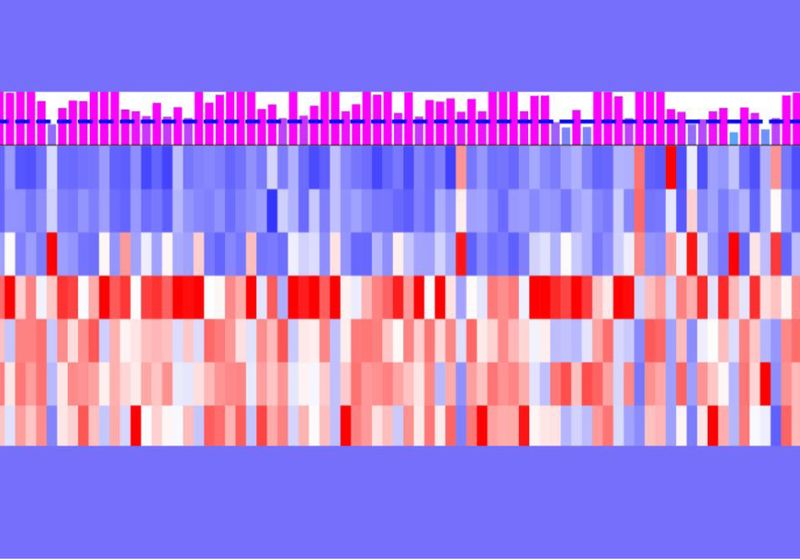
To address Weisel’s question, Zhao and her team expanded on earlier work using single-cell RNA sequencing (scRNA-seq) in Drosophila testes, which had provided high-resolution insight into gene expression patterns.3 In their latest analysis, they applied computational methods to scRNA-seq data from the Fly Cell Atlas and identified nearly 500 de novo gene candidates.4 They found that more than 80 percent of these genes exhibited tissue- or cell-type-specific expression, with the majority localized in the testes and associated with germline cells and sensory neurons—suggesting potential roles for these new genes in reproduction or sensory processing.
The team then asked: Are these de novo genes regulated broadly or are they controlled by some key TFs? From their computational analysis, they found that de novo genes in D. melanogaster were predicted to be regulated by only a few TFs: The top three TFs were Vis, Achi, and Jra (VAJ). “[These TFs] are associated with the meiosis of Drosophila testis, and they basically control roughly a quarter, basically 25 to 28 percent of the de novo gene expression,” explained Zhao. “Meiosis is very important because germ cells need that to produce a sperm for the next generation.”
Zhao and her team investigated the link between de novo gene expression and transcription factors.
Chris Taggart for The Rockefeller University
Then, Zhao and her team generated five fruit fly strains with different copy numbers of vis and achi genes, which are involved in meiotic division and differentiation during spermatogenesis, to see whether vis and achi can affect transcription. Indeed, RNA-seq results showed that Vis and Achi upregulated de novo gene expression compared to other testis-biased genes. This expression also correlated with Vis’ mRNA abundance in a linear manner.
Next, the team sought to see whether these VAJ transcriptional regulations might be linked to de novo gene emergence. They found that these TFs not only regulate and maintain target genes but are also more closely associated with the emergence of younger de novo genes compared to other candidates, suggesting a potential role in their origin.
Caroline Weisman, an evolutionary biologist at Princeton University, who was not involved in the study, noted that the field of evolutionary biology has long leaned toward computational approaches to study genetic questions. “It’s very common to do endless re-identification of putative de novo genes in the genome, sort of like slicing and dicing the data.” She added that the experimental work is the big missing piece, a gap that Zhao’s functional studies help to fill.
Although Weisman is cautious about labeling these genes as truly de novo—preferring terms like “lineage-specific” or “orphan genes”—she acknowledged their significance. “It is very clear to me that whatever they are, they’re very interesting.” She added, “They’re rapidly evolving so fast…[these] genes have always been a class of interesting genes for evolutionary biologists.”
Although many of these de novo genes were specific to the testes, the exact reason for this remains unclear. Some researchers suggest that sexual selection may be a factor. In this study’s findings, Vis and Achi regulate genes important for meiosis in the male germline; however, the exact mechanism driving this process, as Weisman noted, is still an open mystery. Weisman believes there is much more work to be done to better understand this group of mysterious genes.
As Zhao and her team continue to better understand the regulation of de novo genes, she has also turned her sights onto how neighboring genes and regulatory elements coregulate young, new genes.5 Using gene expression patterns and chromatin accessibility data, she and her team can see how genes switch on and become functional. “We haven’t solved the problem yet,” she said. However, the work “gave us some hints about the evolutionary history of gene origination and regulation.”