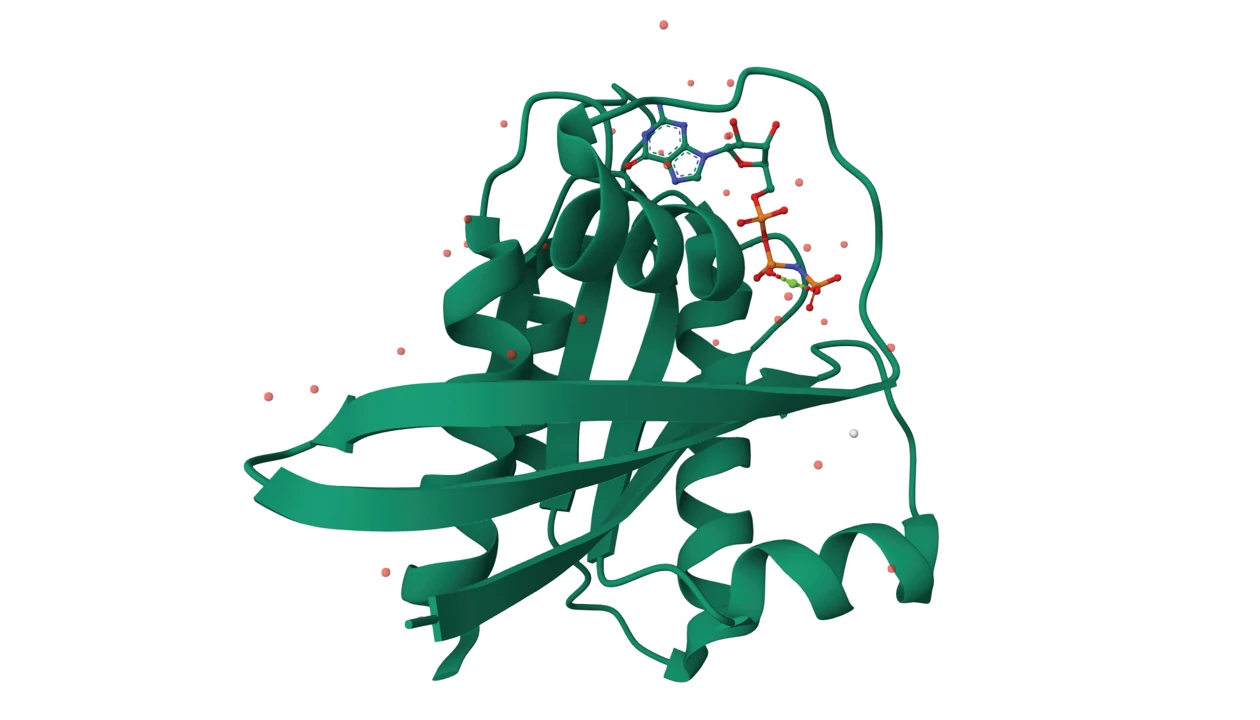
Drug discovery is notoriously slow; finding a single hit can mean screening a million compounds. For stubborn targets like KRas, whose gene is commonly mutated in cancers, the wait can be even longer. Researchers are now working on a way to skip the time-sucking screens done at the bench by using a quantum computer that can explore thousands of drug designs at once.
Christoph Gorgulla, assistant member in the Department of Structural Biology at St. Jude Children’s Research Hospital, described how quantum computing, combined with artificial intelligence, found two candidates that could shut down KRas (Nat. Biotechnol. 2025, DOI: 10.1038/s41587-024-02526-3.) Gorgulla presented the data during a talk on Monday at the American Chemical Society Fall 2025 meeting during a session organized by the Committee of Science.
Unlike classical computers, which use zeros and ones that are arranged to represent instructions, quantum computers use qubits that use a mix of both at the same time. Gorgulla said that this superposition means they can explore many drugs at once—which is much faster than a regular computer, which has to check one drug before moving on to the next.
“I do think it has a big potential for drug discovery. . . . Quantum machine learning could be useful in all sorts of problems where classical machine learning is used,” Gorgulla said.
The workflow Gorgulla described began with a training set—about 1.1 million KRas-targeting molecules assembled from the literature, molecule libraries, and algorithm-generated variants. This dataset taught the quantum model which chemical features tend to appear in promising inhibitors.
Once trained, the quantum model generated thousands of probable patterns for different molecules that fit in the pockets of KRas and stop its function. From there, AI on a classical computer took those and refined them into valid structures. The molecules that were ranked the best by the team’s metrics—potential toxicity, ease of design, and binding affinity—were fed back into the model to improve it.
After several rounds, the algorithm had produced about 1 million potential drug candidates. The team chose the top 15 to synthesize and test in the lab. Measuring how well each one bound KRas and inhibited its activity led to two candidates that stood out: ISM061-018-2 and ISM061-022. These two computer-generated drugs were safe in cells and successful at stopping KRas in cell-based assays, although they may not stand up to other KRas inhibitors out there, Gorgulla said.
“It’s an encouraging early step towards integrating quantum computing into drug discovery,” Angela Wilson, a professor of chemistry at Michigan State University who was not involved in the study, said in an email.
Skeletal structure of KRas inhibitor ISM061-018-2.
Although the system doesn’t quite reach quantum advantage—proof that no classical method could match this—with further development, it could be a faster way for drug discovery, Gorgulla said.
However, Gorgulla said that for quantum computers to beat classical ones, researchers will have to find ways to increase processing power, which could be 4 or 5 years away.
Chemical & Engineering News
ISSN 0009-2347
Copyright ©
2025 American Chemical Society