GenMasterTable is a standalone, cross-platform desktop application developed in Python to facilitate scalable genomic data analysis. It integrates several high-performance libraries, including Pandas for tabular data manipulation, NumPy for numerical operations, PyVCF for variant call format (VCF) parsing, and tqdm for progress monitoring. The graphical user interface (GUI) is built using Tkinter and incorporates an embedded pandastable table viewer, enabling users to interactively explore, filter, and analyze both VCF and tabular genomic datasets.
Application architecture and GUI design
The GUI adopts a vertically split PanedWindow layout that separates data visualization from user controls. The upper panel displays a scrollable and sortable variant table, while the lower panel provides access to file operations, filtering tools, and export options. This architecture ensures a streamlined, intuitive workflow for navigating and manipulating large datasets.
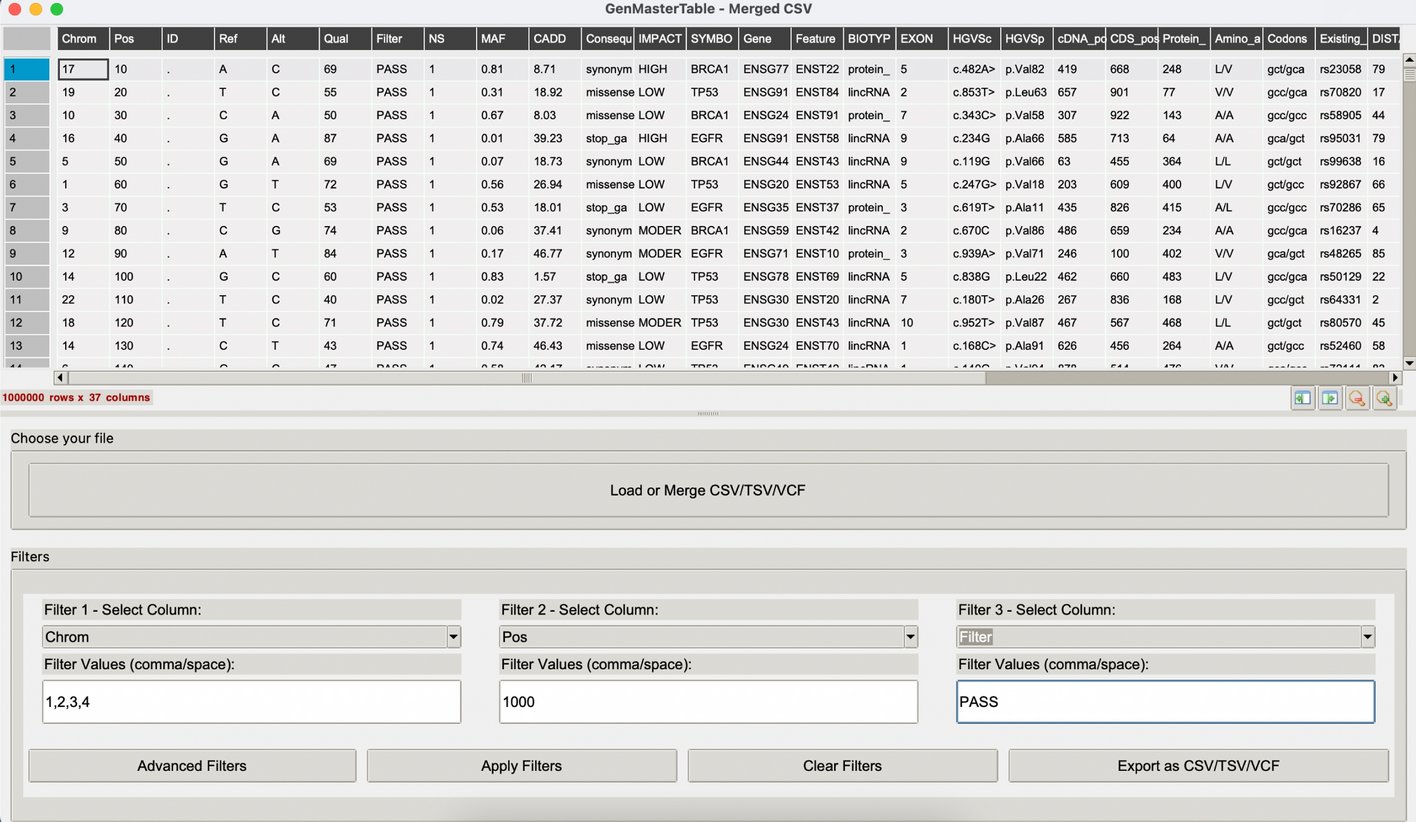
GenMasterTable supports the loading and concatenation of multiple annotated files in CSV, TSV, or VCF format. Files selected in a single session are concatenated into a unified dataset. To retain data provenance, each record is tagged with a File_Name field that reflects its original source. Internally, all parsed content is consolidated into a unified Pandas DataFrame, which serves as the foundation for downstream filtering, transformation, and visualization. Figure 1 presents a demonstration of GenMasterTable’s GUI, applied to an artificially generated test dataset.
GenMasterTable graphical user interface for variant filtering and exploration. Screenshot of the GenMasterTable desktop application, showing the main interface for loading and interacting with annotated variant datasets. The top panel displays the merged and parsed VCF data in a tabular format with sortable columns. The lower panel provides user-friendly filtering options, allowing intuitive column-based filtering using dropdowns and value inputs without requiring programming. Users can load or merge multiple CSV/TSV/VCF files and export filtered results directly
Data parsing and chunked processing
To ensure scalability for large genomic datasets, GenMasterTable employs a chunked data loading strategy that minimizes memory usage. CSV and TSV files are read in successive segments, allowing the application to efficiently handle files containing millions of rows without exhausting system resources.
For VCF files, GenMasterTable utilizes the PyVCF library to parse records in batches. For each variant, key attributes—including chromosome (CHROM), position (POS), reference and alternate alleles (REF, ALT), quality scores (QUAL), filter status (FILTER), INFO fields, and genotype calls (GT)—are systematically extracted and structured. The INFO column is parsed by splitting each key-value pair using the field delimiter defined in the VCF specification, and the resulting annotations are mapped to individual columns using the corresponding VCF header definitions.
All parsed records are concatenated into a single unified Pandas DataFrame, supporting efficient querying, visualization, and comparative analysis across multiple samples or conditions.
Filtering system
GenMasterTable implements a dual-mode filtering system to support both rapid querying and complex data subsetting. The basic filtering mode provides three independent filter fields, each allowing users to select a column and specify inclusion criteria using comma- or space-separated values. This mode is ideal for quickly filtering categorical or numerical variables in isolation.
The advanced filtering mode is accessible via a dedicated button that launches a dynamic pop-up interface. Within this interface, users can define multiple logical filtering rules, where each rule consists of a selected column, a comparison operator (e.g., “equals,” “contains,” “ ≥,” or “is not empty”), and an optional input value. The interface intelligently adapts to the selected operator, automatically showing or hiding input fields as appropriate to reduce user error.
This flexible design enables users to build complex, multi-condition queries across several columns, making GenMasterTable suitable for both exploratory data inspection and hypothesis-driven variant selection.
Interactive data operations
To support exploratory data analysis, GenMasterTable offers a suite of interactive features. Users can sort columns based on gene names, chromosomal positions, allelic depth, quality scores, or any custom annotation. Columns may be deleted from the display, and a type assignment function allows users to explicitly convert column types to numeric, categorical, or text formats, ensuring compatibility with the filtering engine.
The application also includes a counting analysis function that computes value counts across selected columns, aiding in the identification of recurrent variants or annotations across the dataset. All operations are tightly synchronized with the internal DataFrame and the GUI, ensuring that user actions are immediately reflected across the interface without inconsistencies.
Comprehensive usage examples and a step-by-step walkthrough are available in the user manual on the GenMasterTable GitHub repository, which also includes a demonstration using synthetic data.
Informative message handling
Throughout the application, GenMasterTable employs a consistent and user-friendly system of pop-up message boxes to provide feedback and prevent critical errors. Message boxes are used to notify users of successful operations, such as file loading, filtering, or export completion, as well as to warn users of missing inputs, type mismatches, or other potential issues. Error messages are automatically triggered in scenarios such as attempting to export an incomplete VCF, selecting unsupported file types, or applying incompatible filter criteria. These message dialogs improve robustness and usability, especially for non-programming users.
Data export and VCF reconstruction
Processed datasets can be exported in CSV, TSV, or VCF format through an integrated export menu accessed via a dedicated button. For CSV and TSV outputs, the data are written directly using Pandas’ to_csv function with appropriate delimiters.
VCF export is supported only when the input includes at least one VCF file. In such cases, GenMasterTable reconstructs valid VCF records using the original PyVCF header objects retained from the initial file loading process. Each variant record is reassembled with the required fields—CHROM, POS, REF, and ALT—and includes relevant metadata such as quality scores, filters, and INFO annotations. Export is permitted only if all mandatory fields are present in the filtered dataset. If any required fields have been deleted or renamed during analysis, the system automatically alerts the user and prevents invalid export.
During export, variants are grouped by their original input file to maintain header integrity and preserve metadata structure. This ensures that output files conform to standard VCF specifications and remain suitable for downstream use, including reanalysis or database submission.
Platform compatibility and deployment
GenMasterTable is compatible with Windows, macOS, and Linux operating systems. The application is distributed as a self-contained executable using PyInstaller, producing.exe files for Windows,.app bundles for macOS and.ELF (Executable and Linkable Format) for Linux. Installation requires no additional dependencies or administrative privileges. GenMasterTable is designed to function entirely offline, making it ideal for use in secure computing environments such as clinical research settings or hospital systems where data privacy and local execution are essential.