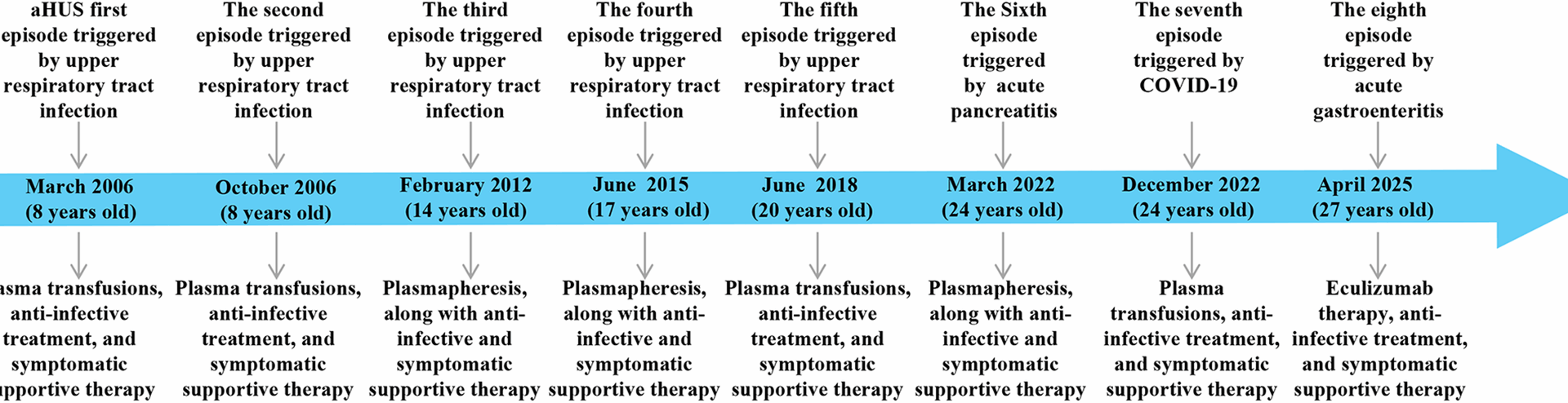
The patient is a 27-year-old male who was admitted to our hospital on March 24, 2022, owing to “diagnosed aHUS for 16 years, abdominal pain for 2 days, and brown urine for 2 hours”. A timeline of the patient’s medical history is shown in Fig. 1.
The patient first presented to Shanghai Children’s Hospital affiliated with Fudan University on March 6, 2006, due to fever accompanied by cough, sputum production, jaundice, and gross hematuria. Laboratory tests indicated renal function abnormalities, anemia, and thrombocytopenia. He was diagnosed with “aHUS and bronchopneumonia”. Treatment included antibiotics such as cefotaxime and penicillin, plasma transfusions and urine alkalinization. The patient’s renal function, hemoglobin levels, and platelet counts returned to normal, and he was discharged. The patient returned to Fudan University Children’s Hospital on four occasions (October 28, 2006; February 10, 2012; June 24, 2015; and June 6, 2018) due to fever, cough, sputum production, and gross hematuria, with all cases considered aHUS triggered by acute upper respiratory infections. For the episodes in February 2012 and June 2015, the patient underwent plasmapheresis, along with anti-infective and symptomatic supportive therapy. For the other two episodes, the patient received plasma transfusions, anti-infective treatment, and supportive therapy. All four episodes ultimately fully resolved, and renal function and complete blood count tests remained normal at follow-up visits between episodes. Two days prior to this latest admission, the patient experienced upper abdominal pain without any obvious trigger, accompanied by nausea and dry heaving, with no diarrhea or melena. The pain gradually worsened, and two hours before admission, the patient developed tea-colored urine along with lumbar pain, leading to a visit to our hospital’s emergency department. The examination results were as follows: urinalysis (2022-03-24, Emergency): urine protein 2+, urine occult blood 3+; blood amylase, 866 U/L; urine amylase, 1831 U/L; blood creatinine, 281.4 µmol/L; lactate dehydrogenase, 2454.4 U/L; schistocytes, 10%; and complete blood count: white blood cell count, 11.79 × 109/L; neutrophil percentage, 80.8%; hemoglobin, 142 g/L; and platelet count, 78 × 109/L. On the basis of these findings, aHUS was suspected, and the patient was admitted to the nephrology ward. The patient’s medical history included hypertension for 3 years and a history of plasma and red blood cell transfusions. The patient’s family history was as follows: his parents were affected by blood (cousins), and the patient’s grandmother, mother, and cousin had a history of anemia with specific causes unknown. No other obvious familial or hereditary diseases were noted. The physical examination results were as follows: temperature, 36.5 °C; pulse, 74 beats/min; respiration, 19 breaths/min; blood pressure, 124/80 mmHg; and weight, 90 kg. The patient was alert and in good spirits. Heart, lung, and abdominal examinations were normal, and there was no swelling in the lower limbs. Pathological signs were negative. Key laboratory abnormalities upon admission are summarized in Table 1. Additional investigations, including antinuclear antibody (ANA), antineutrophil cytoplasmic antibodies (ANCA), anti-glomerular basement membrane antibody (anti-GBM), serum glucose, electrolytes (sodium, potassium, chloride, calcium), coagulation profile, and serological markers for hepatitis and HIV, as well as an electrocardiogram and chest computed tomography, yielded unremarkable results.
Patient history timeline. In the diagram, the blue thick arrows represent the treatment timeline, and the grey arrows represent the key time points. The upper part of the arrows indicates recurrence triggers: clinical events that precipitated aHUS episodes, while the lower part indicates treatment measures
Table 1 Laboratory data at admission
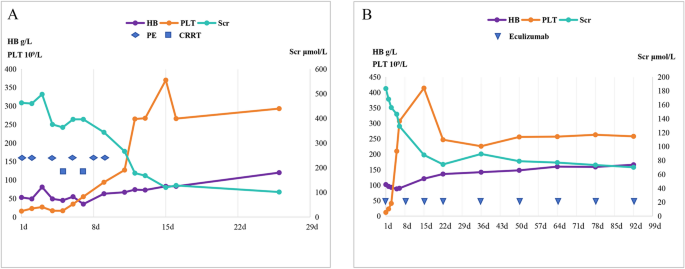
Diagnosis and treatment process The patient was admitted with upper abdominal pain and markedly elevated amylase levels (serum: 866 U/L; urine: 1831 U/L). Initial laboratory results revealed a hemoglobin level of 160 g/L, a platelet count of 242 × 109/L, and a creatinine level of 187 µmol/L. On the 3rd day, a follow-up test revealed that the hemoglobin level decreased to 53 g/L, the platelet count decreased to 16 × 109/L, and the creatinine level increased to 462.9 µmol/L. The patient experienced progressively worsening hemolytic anemia and deteriorating kidney function. Considering the patient’s medical history and the results of relevant tests conducted after admission, a diagnosis of aHUS and acute pancreatitis was established. During hospitalization, the patient underwent 6 plasma exchanges and 2 continuous renal replacement therapy (CRRT) sessions. Supportive care included antibiotic treatment, red blood cell transfusions, and symptomatic management. On April 9, 2022, follow-up tests revealed that total bilirubin was 10.5 µmol/L, direct bilirubin was 3.7 µmol/L, creatinine decreased to 128.0 µmol/L, hemoglobin improved to 83 g/L, and the platelet count returned to 266 × 109/L. The patient’s condition improved significantly, and he was discharged. After discharge, the patient was regularly followed up in our department every month for liver and kidney function tests, complete blood count, all of which were normal (Fig. 2A). On December 25, 2022, the patient visited Eastern Theater General Hospital again due to fever, cough with sputum, and gross hematuria. A routine blood test revealed a white blood cell count of 11.49 × 109/L, a neutrophil percentage of 82%, a hemoglobin level of 60 g/L, a platelet count of 18 × 109/L, and a serum creatinine level of 231 µmol/L. Concurrently, the patient tested positive for SARS-CoV-2 via nucleic acid amplification. Based on the clinical presentation and laboratory findings, a diagnosis of aHUS secondary to COVID-19 infection was suspected. Owing to insufficient blood supply, plasma exchange was not performed. During hospitalization, symptomatic treatments, including infection control and plasma transfusion, were administered, and the patient’s condition completely improved before discharge. However, on April 22, 2025, the patient was readmitted due to a relapse of aHUS triggered by acute gastroenteritis. Admission laboratories revealed hemoglobin 102 g/L, platelets 12 × 109/L, and serum creatinine 183 µmol/L. Notably, eculizumab had been included in China’s National Reimbursement Drug List (NRDL) for aHUS on January 1, 2024, substantially reducing financial barriers to treatment. Given this improved accessibility, eculizumab was recommended as first-line therapy. Due to critical disease progression requiring immediate intervention, timely meningococcal vaccination could not be administered. After obtaining written informed consent from the patient and his parents, eculizumab therapy was initiated within 24 h of relapse onset, with concurrent antibiotic prophylaxis using piperacillin-tazobactam. Meningococcal vaccination was subsequently completed within two weeks of initial eculizumab administration. Treatment outcomes demonstrated a rapid increase in platelet counts within 24 h of initiation, achieving normalization within one week. Hemoglobin levels and renal function also showed improvement beginning in the first week, reaching complete normalization within two weeks. As of the last follow-up in July 2025, hepatic and renal function along with complete blood count parameters remained within normal ranges, while the patient continued on biweekly eculizumab maintenance therapy (Fig. 2B).
Treatment and follow-up process. (A) Clinical course during the seventh episode (March 2022). “1d” indicates initiation of plasma exchange (PE; dark blue rhombuses). The patient received 6 PE sessions and 2 continuous renal replacement therapy (CRRT; dark blue squares) sessions. (B) Clinical course during the eighth episode (April 2025). “1d” indicates initiation of eculizumab therapy (dark blue triangles) with the following regimen: 900 mg weekly on Days 1, 8, 15, and 22, 1200 mg on Day 36, then 1200 mg every 14 days thereafter
Line indicators: Blue, serum creatinine (Scr); purple, hemoglobin (HB); orange, platelet count (PLT). Intervention markers: Dark blue rhombuses, PE; Dark blue squares, CRRT; Dark blue triangles, eculizumab therapy.
We performed extensive diagnostic testing to identify the cause of our patient’s HUS. The study was performed in accordance with the Declaration of Helsinki and was approved by the ethics committee of the Fuyang People’s Hospital Affiliated with Anhui Medical University (Number: 2025-19), and all participants signed written informed consent forms.
Genetic investigation methodWhole-exome sequencing (WES)
Relevant data from the patient, including medical history and family history, physical examination results upon admission, and auxiliary examination results, were collected. A GenCap® Whole Exome Gene Capture Probe V6.0 (Beijing Maikeno Gene Technology Co., Ltd.) was used to perform hybrid capture of the patient’s whole exome. The target area included 23,000 genes, with a total length of 51 Mb. High-throughput sequencing was conducted on the captured library, achieving an average sequencing depth of 134.03X. Candidate variant gene loci were identified through bioinformatics analysis.
Sanger sequencing family verification
Based on the high-throughput sequencing results, primers were designed to amplify the target fragments. Genomic DNA was extracted from 50 to 100 µL of EDTA-anticoagulated whole blood using a commercial kit (G3633; Wuhan Servicebio Technology Co., Ltd.). Using these primers, target fragments were amplified by polymerase chain reaction (PCR) in 50 µL reactions containing 25 µL of 2× Fast Pfus PCR Master Mix (G3305; Wuhan Servicebio Technology Co., Ltd.), 1.5 µL of each forward and reverse primer (10 µM; sequences listed in Supplementary Table 1), 2 µL of DNA template, and nuclease-free water to a final volume of 50 µL. The thermocycling conditions consisted of an initial denaturation at 98 °C for 2 min, followed by 30 cycles of denaturation at 98 °C for 20 s, annealing at 55 °C for 20 s, and extension at 72 °C for 10 s, with a final extension at 72 °C for 5 min. Reactions were then held at 16 °C. PCR products were verified by 2% agarose gel electrophoresis and were bidirectionally sequenced on an ABI 3730xl platform (Applied Biosystems, USA) by Wuhan Servicebio Technology Co., Ltd. Sequence chromatograms were analyzed using Chromas v2.6.6 (Technelysium Pty Ltd.) to confirm pathogenic variants identified by high-throughput sequencing.
Detection of CD46 mRNA expression by fluorescence quantitative PCR
A total of 5 ml of peripheral venous blood was collected from the patient, the patient’s mother, and healthy control. Total RNA was extracted using a commercial kit (G3013; Wuhan Servicebio Technology Co., Ltd.) and its concentration/purity assessed spectrophotometrically. cDNA was synthesized from 500 ng − 1 µg of total RNA using the SweScript All-in-One RT SuperMix for qPCR (One-Step gDNA Remover) (G3337; Wuhan Servicebio Technology Co., Ltd.) in a 20 µl reaction volume according to the manufacturer’s instructions. Reverse transcription was performed at 25 °C for 5 min, 42 °C for 30 min, and 85 °C for 5 s using a thermal cycler (ETC811; Beijing Dongsheng Innovation Biotechnology Co., Ltd.). SYBR Green qPCR was performed using the 2× Universal Blue SYBR Green qPCR Master Mix (G3326; Wuhan Servicebio Technology Co., Ltd.) and 0.25 µM (final concentration) of each specific primer (sequences listed in Supplementary Table 2) in a 15 µl reaction volume: 7.5 µl Master Mix, 0.75 µl of each primer (2.5 µM stock), 2 µl cDNA, and nuclease-free water to 15 µl. The amplification protocol consisted of: 95 °C for 30 s; 40 cycles of 95 °C for 15 s and 60 °C for 30 s (annealing/extension); followed by a melting curve analysis (65 °C to 95 °C, 0.5 °C increments with a 5-sec hold/fluorescence acquisition at each step). Three technical replicates were performed per sample. Relative CD46 mRNA expression, normalized to GAPDH, was calculated via the 2−ΔΔCT method.
Detection of the CD46 protein expression level in the plasma via enzyme-linked immunosorbent assay (ELISA)
A human CD46 ELISA detection kit with a kit specification of 48T was used. The testing instrument used was an enzyme-linked immunosorbent assay analyzer (Rayto RT-6100) (Wuhan Servicebio Technology Co., Ltd.). The testing methods and operational steps were performed according to the instructions provided in the manual.
Genetic investigation resultsWES genetic sequencing results
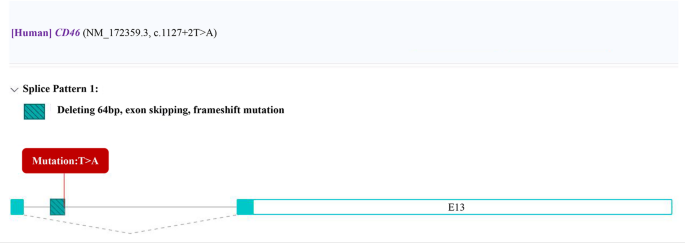
High-throughput WES revealed a homozygous mutation in the first nucleotide of intron 12 of the CD46 gene in the proband (NM_172359.3, c.1127 + 2T > A, gene location 207785684). According to the prediction results from the Rare Disease Data Center (RDDC) RNA splicer algorithms (https://rddc.tsinghua-gd.org/) [15], this alteration in the sequence may lead to a change in the reading frame, exon skipping, and deletion of a 64 bp fragment, causing a frameshift variant (Fig. 3). Consequently, it induces alternative splicing, affects protein coding, and may lead to a splicing-related disease [16]. According to ACMG guidelines [17], this variant is classified as a pathogenic variant (PVS1 + PM2_Supporting + PM3_ Supporting): this variant is a null variant (splicing mutation) that may result in loss of gene function (PVS1); the frequency in the general population database is zero (PM2); and the variant is a homozygous variant in a recessive inheritance gene (PM3).
Prediction results of RDDC RNA splicer algorithms: The alteration in the sequence may lead to a change in the reading frame, exon skipping, and deletion of a 64 bp fragment, causing a frameshift variant
Sanger sequencing validation results
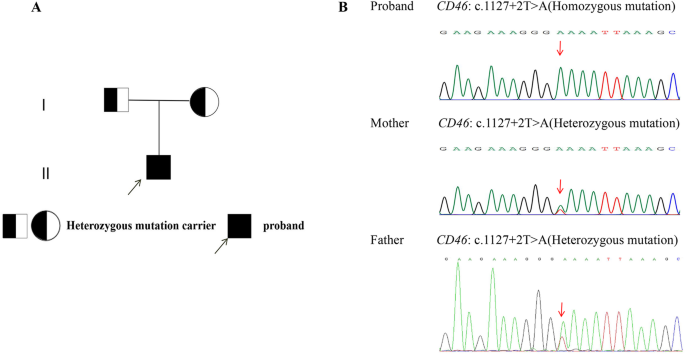
The proband harbors a homozygous mutation in the first nucleotide of intron 12 of the CD46 gene (c.1127 + 2T > A), whereas the parents harbor a heterozygous mutation (c.1127 + 2T > A). The pedigree of the proband’s family and Sanger sequencing chromatogram of CD46 are shown in Fig. 4.
Pedigree and Sanger sequencing of the pathogenic CD46 splice-site variant (c.1127 + 2T > A). (A) Pedigree showing autosomal recessive inheritance: proband homozygous for c.1127 + 2T > A, parents heterozygous. (B) Sanger chromatograms: Proband (Homozygous): T > A substitution at intron 12 donor site (+ 2 position, NM_172359.3). Parents (Heterozygous): Co-occurrence of wild-type T and mutant A peaks. Functional impact: Disrupts canonical splicing (GT→GA), predicted to cause exon skipping, 64-bp deletion, and frameshift leading to loss-of-function. Classified as pathogenic (PVS1 + PM2_Supporting + PM3_Supporting) per ACMG guidelines
Detection of peripheral blood mRNA via fluorescence PCR
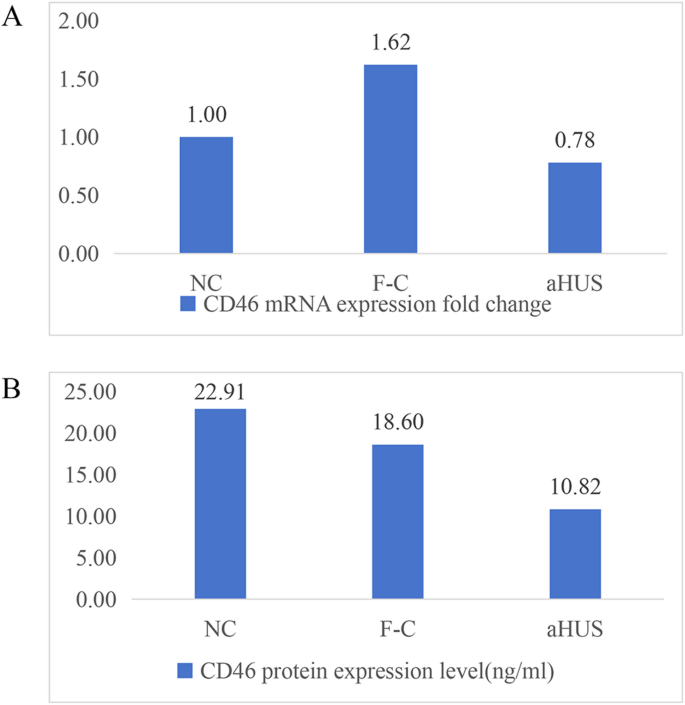
The fold change in CD46 mRNA expression in peripheral whole blood was 0.78 for the proband, 1.62 for the mother, and 1 for the healthy control (Fig. 5A).
Plasma CD46 protein expression levels detected by ELISA
The CD46 protein level in the peripheral blood of the proband was 10.82 ng/ml, that in the mother was 18.06 ng/ml, and that in the healthy control was 22.91 ng/ml (Fig. 5B).
Peripheral blood mRNA results and plasma CD46 protein expression levels. (A) Fluorescence PCR detection of peripheral blood mRNA in the proband (aHUS), proband’s mother (F-C), and healthy control (NC); (B) Plasma CD46 protein expression levels in the proband (aHUS), proband’s mother (F-C), and healthy control (NC)