Among the most important groups of genes that coordinate plant development are those that regulate phytohormone homeostasis and content. The genes belong to gene families and are expressed specifically in selected organs or spatio-temporally through developing plants. Prior recognition of their patterns of expression in different organs of developing plants is an initial step in characterizing their function.
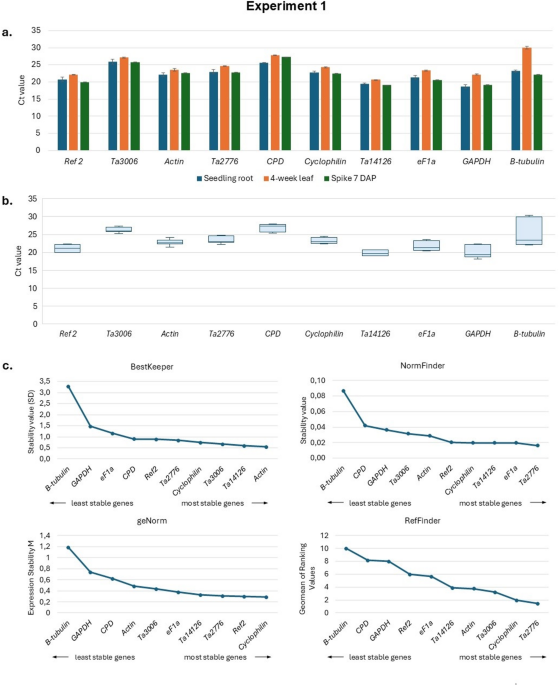
Quantitative, real-time PCR (qPCR) is among the most precise and reliable techniques for gene expression analysis. However, selecting and validating stable reference genes is crucial for accurate normalization, especially in complex allohexaploid species such as Triticum aestivum (common wheat). In our unpublished preliminary research, we identified ADP-ribosylation factor (Ref 2) as the most suitable reference gene for studying the expression of spatio-temporally regulated gene families. Ref 2 has been successfully used in previous studies on the expression patterns of TaCKX genes13,41,42and TaNAC genes43. However, some researchers have opted to use two or even more reference genes to obtain more comparable results27. Therefore, before studying the next family of phytohormone-regulating genes, we conducted research on ten candidate reference genes for their stability of expression in various tissues and developmental stages of wheat, and validated them through rigorous analysis using multiple statistical tools (BestKeeper24NormFinder23geNorm20 and RefFinder25,46).
Identification of stable reference genes
A three-step analysis was conducted to limit the number of experimental trials. In Experiment 1, we evaluated ten candidate reference genes using three very diverse tissues/organs collected at different developmental stages of plant growth. The goal was to pre-select the most suitable and exclude the most unstable reference genes. These ten candidate reference genes were assessed in three tissues of the Ostka cultivar, revealing considerable variation in expression stability. Genes such as β-tubulin, CPD, and GAPDH exhibited high variability and were deemed unsuitable for normalization. Conversely, Ref 2, Ta2776, Cyclophilin, Ta3006, and Ta14126 consistently ranked as the most stable across all software analyses. Similarly, GAPDH was identified as the least stable reference gene during endosperm development in wheat, as reported by Mu et al.28.
Expanding the analysis in Experiment 2, six selected genes were evaluated in five tissues, further validating Ref 2, Ta3006, Ta2776, and Cyclophilin as robust reference genes. The exclusion of CPD and Actin from this group underscores the importance of tissue-specific testing, as these genes, although widely used in other contexts, proved less stable under the conditions tested here.
Part of these findings is consistent with previous studies highlighting the reliability of Ref 2/Ta2291, Ta2776 and Cyclophilin as the most stable reference genes for normalizing gene expression in different tissues and development stages of wheat21. In their research, these genes outperformed all traditional housekeeping genes such as actin and α-tubulin, β-tubulin, Ubiquitin, and GAPDH. Conversely, the authors did not find Cyclophilin as stable as in our research. However, the same reference gene was among the two best for studying wheat flag leaf expression in three cultivars grown under different farming conditions30. Similarly, Ref 2 was validated as the most stable gene to study grain filling in wheat29. The reliability of this reference gene was demonstrated by monitoring the expression dynamics of three NAM genes (TaNAM-A1, TaNAM-B1, and TaNAM-B2) in flag leaves or the 10 samples tested. Their results suggest that this single reference gene, identified by geNormPlus, is optimal for use in this experiment. The Elongation factor 1 alpha-subunit 2 was the second in ranking, though noticeably less stable than Ref 2. Glyceraldehyde-3-phosphate dehydrogenase, translation elongation factor EF-1alpha (TEF1), CPD, actin and tubulin gene families were also used to study wheat meiosis31 and in other studies47.
Several reference genes that we used to study expression of spatio-temporally regulated genes in different organs of developing wheat plants have also been used in stress response studies47. In studies of gene expression under biotic or abiotic conditions, actin was the best candidate among housekeeping genes in common wheat seedlings under short-term drought stress34. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was ranked among the three most stable internal control genes for studying viral infections in cereals35. On the other hand, elongation factor-1 alpha for barley and oat samples, and α-tubulin for wheat samples, were consistently ranked as the less reliable controls. In durum wheat (Triticum durum L.) under drought stress, the most stable reference genes were GAPDH, ubiquitin and β-tubulin2, whereas under salinity stress conditions, the most stable reference genes were eukaryotic elongation factor 1-α, glyceraldehyde-3 phosphate and actin33. Scholtz and Visser36 documented that there is a need for validation of reference genes for the analysis of the expression of each different plant-pathogen interaction.
Several reference genes tested by us were also validated in other cereal genomes such as hexaploid oat (Avena sativa L.)22. The most stable for all samples starting from seedling shoots and roots to developing seeds and endosperms was EIF4A (Eukaryotic initiation factor 4 A-3). The same reference gene EF1A (elongation factor 1‑alpha) was among the two most stable in in Avena sativa during compatible and incompatible interactions with two different pathotypes of Puccinia coronata f. sp. Avenae37. In the same system, Cyclophilin was shown to be the worst candidate. EF1A (elongation factor 1‑alpha) was also one of the three most appropriate reference genes in studies of compatible and incompatible interactions with Puccinia graminis in the same cereal species38. These differences in the stability of reference genes can be dependent on experimental system and conditions, as well as species specificity and their interaction with pathogens. As documented by Yang et al.22 in oat, the feasibility of reference genes for qPCR in polyploid species is influenced by the number of copies of these genes.
Validation across cultivars and tissues
In the subsequent experiment comparing two wheat cultivars (Kontesa and Ostka), Ref 2 and Ta3006 demonstrated consistent expression across twelve tissues, further supporting their suitability as reference genes. This consistency between cultivars enhances their utility in broader genetic and physiological studies of wheat, addressing concerns about genotype-specific variability. The Ct values for Ref 2 and Ta3006 in all samples ranged between 20 and 24, and 25 and 30, respectively, reflecting uniform expression levels regardless of tissue type or developmental stage.
Application to normalization of target genes
The utility of these reference genes was further demonstrated in normalization of TaIPT1 and TaIPT5 gene expression. TaIPT1, which is specifically expressed in developing spikes, did not show significant differences between absolute and normalized expression values when either Ref 2 or Ta3006 was used. This highlights the precision and reliability of these reference genes in quantifying tissue-specific expression patterns. Similarly, for TaIPT5, a broadly expressed gene, normalization with Ref 2, Ta3006, or their combination yielded consistent results, emphasizing their robustness even in complex expression analyses.
Comparison of single and dual reference genes
Although normalization of IPT1 and IPT5 using single or dual reference genes yielded similar trends, the use of two reference genes can enhance precision and reliability, especially in experiments involving multiple tissues or stress conditions. This observation aligns with earlier recommendations for using multiple reference genes in gene expression studies19,20,21. However, our findings are also consistent with a study on grain filling in wheat, where Ref 2 was identified as an optimal single reference gene29.
Implications, limitations and future directions
The validated reference genes identified in this study, particularly Ref 2, Ta3006, Ta2776, and Cyclophilin, provide a robust foundation for RT-qPCR-based expression analyses in wheat. Their consistent performance across tissues, developmental stages, and cultivars underscores their utility in diverse experimental contexts.
This study was conducted under standard growth conditions, using two cultivars, twelve diverse tissues/organs collected at different developmental stages, and ten candidate reference genes. Although two wheat cultivars were analyzed, the findings may not fully represent the genetic diversity of wheat or related Triticeae species. Therefore validation of reference genes in additional wheat varieties and related Triticeae species could further expand their applicability. Further research should also investigate their stability under various abiotic and biotic stress conditions to extend their relevance to studies of stress responses in wheat.
In conclusion, this study identifies and validates four reliable reference genes for RT-qPCR normalization in wheat: Ref 2, Ta3006, Ta2776, and Cyclophilin, facilitating accurate expression analysis of developmentally regulated genes. Among them Ref2 and Ta3006 had comparable expression levels in different genotypes across the tissues tested, and each is sufficient for normalization of developmentally regulated genes in wheat. Through rigorous analysis using multiple statistical tools (BestKeeper, NormFinder, geNorm, and RefFinder), our findings contribute significantly to optimizing RT-qPCR normalization in wheat. These findings will help researchers characterize gene functions, advancing molecular breeding efforts, and ultimately improve wheat productivity and resilience.