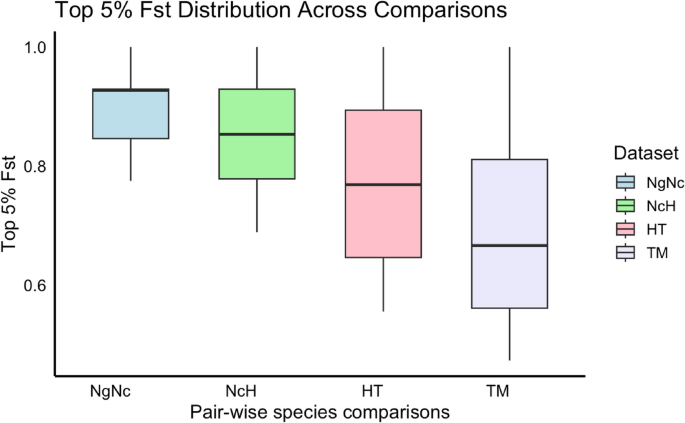
Fst distribution
In total, we identified over 200,000 SNPs in each pairwise comparison of Sulawesi macaque species (Supplemental data). The total number of SNPs ranged from 215,435 in the comparison of NgNc to 411,879 in the comparison of TM (Table 1). Notably, the highest number of SNPs was detected in the comparison of TM, which showed relatively low genetic differentiation with a mean Fst of 0.104. The mean Fst values for other comparisons were 0.108 for HT and 0.148 for NcH. In contrast, the NgNc comparison exhibited higher genetic differentiation, with a mean Fst of 0.179. The Fst distributions for all four comparisons were strongly skewed, with approximately 40–60% of SNPs having an Fst below 0.05 (Fig. 2).
Table 1 An overview of the SNP calling results and the top 5% Fst value SNPs identified in the four pairwise comparisons: M. nigra/M. nigrescens (NgNc), M. nigrescens/M. hecki (NcH), M. hecki/M. tonkeana (HT) and M. tonkeana/M. maurus (TM)Fig. 2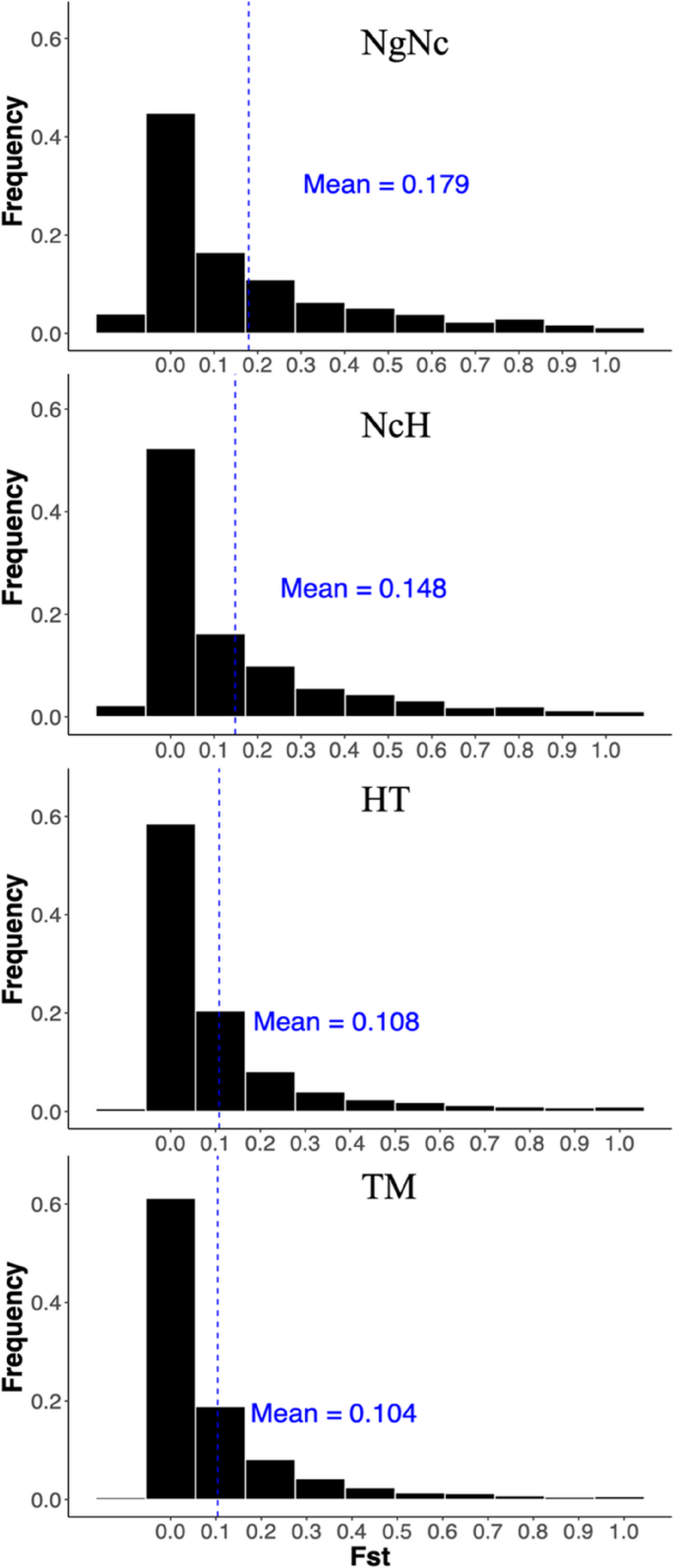
Distribution of Fst values in all four pairwise comparisons. Histogram showing Fst distribution between M. nigra/M. nigrescens (NgNc), M. nigrescens/M. hecki (NcH), M. hecki/M. tonkeana (HT) and M. tonkeana/M. maurus (TM). Fst distributions for all four comparisons were strongly skewed with approximately 40–60% of SNPs having an Fst under 0.05. Mean of Fst value was highlighted in blue
Identification of highly divergent genomic regions
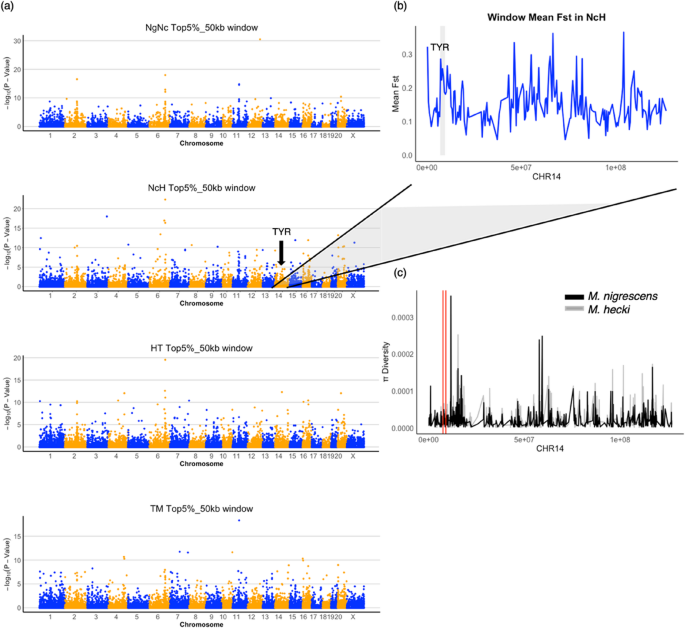
To determine the genomic regions under potential selection, we identified putative targets enriched with the SNPs within the top 5% of Fst value in each comparison. The mean Fst values for the top 5% SNPs in comparisons were 0.899 (SD = 0.074) in NgNc, 0.857 (SD = 0.100) in NcH, 0.771 (SD = 0.138) in HT, and 0.699 (SD = 0.162) in TM, respectively. Furthermore, candidate regions were identified using Fisher’s exact test with a significance threshold of P −5 (Fig. 3a), resulted in 272, 229, 192, 206 candidate genes in NgNc, NcH, HT, and TM, respectively. Approximately 31.41–32.94% of these highly divergent SNPs were located in exonic regions. In total, 550 missense SNPs were identified across four comparisons (Table S2).
The distribution of the top 5% SNPs Fst values across all four pairwise species comparisons: M. nigra/M. nigrescens (NgNc), M. nigrescens/M. hecki (NcH), M. hecki/M. tonkeana (HT) and M. tonkeana/M. maurus (TM)
Functional enrichment of candidate genes
To explore the possible functional implications of the candidate genes, we performed gene ontology (GO) and pathways enrichment analysis. The top 20 most significantly enriched clusters for each comparison are summarized in Table S3. In NgNc, overrepresented GO terms included “mRNA processing” and “homophilic cell adhesion via plasma membrane adhesion molecules,” while pathways such as “Signaling by Rho GTPases” and “Viral Infection Pathways” were enriched. Similarly, NcH candidate genes were enriched in pathways like “TGF-beta signaling pathway” and GO terms including “intracellular protein transport” and “mitotic cell cycle process.” Notably, 26 genes (HMGCR, HYAL1, PPP1CC, ROM1, TP53, TYR, HYAL3, MAP4K3, SLC24A1, TIPIN, LRIT3, CD44, NABP1, KDM5B, CD14, CYP1B1, STAT1, B4GALT3, B4GAT1, GALK1, NT5C2, SAMHD1, AAAS, PIK3CA, GANAB, PGGHG) in NcH were associated with “response to light stimulus,” with six them (HYAL1, TP53, TYR, HYAL3, MAP4K3, TIPIN) further linked to “response to UV,” highlighting their role in pigmentation regulation.
Variants in TYR and pigmentation genes
Notably, the TYR (Tyrosinase) gene, known for its critical role in triggering the first and rate-limiting step in melanin biosynthesis, exhibited significant genetic variation in Sulawesi macaques. Among the top5% SNP, we detected 10 variants located in TYR, comprising 5 synonymous SNPs, 3 intron SNPs and 2 missense SNPs. The genomic windows containing the TYR gene showed high level of differentiation in the NcH comparison (Fig. 4b) and exhibited low nucleotide diversity in both M. nigrescens and M. hecki (Fig. 4c). Both analyses were performed using non-overlapping 50-kb windows.
Collapsing analysis identifies TYR as a highly divergent gene. a Manhattan plots of the population differentiation in 50-kb nonoverlapping windows across all four pairwise species comparisons: M. nigra/M. nigrescens (NgNc), M. nigrescens/M. hecki (NcH), M. hecki/M. tonkeana (HT) and M. tonkeana/M. maurus (TM). b Manhattan plot of 50-kb mean Fst across Chromosome 14 in NcH comparison. The dashed block highlights the neighboring windows and TYR window. c Nucleotide diversity for 50-kb nonoverlapping windows across Chromosome 14 in M. nigrescens and M. hecki. The red lines highlight the neighboring windows and TYR window
Notably, four of these SNPs were located in the first exon (Table 2). Furthermore, these four SNPs were shared between M. nigrescens and M. nigra, distinguishing them from the other Sulawesi macaque species (Fig. 5a).
Table 2 Diverged SNPs in TYR gene in all four comparisons. The values represent site-specific Fst estimates for the comparisons: M. nigra/M. nigrescens (NgNc), M. nigrescens/M. hecki (NcH), M. hecki/M. tonkeana (HT) and M. tonkeana/M. maurus (TM). Each row represents a SNP detected as top5% Fst value in one or more comparisons. SNP: single nucleotide polymorphismFig. 5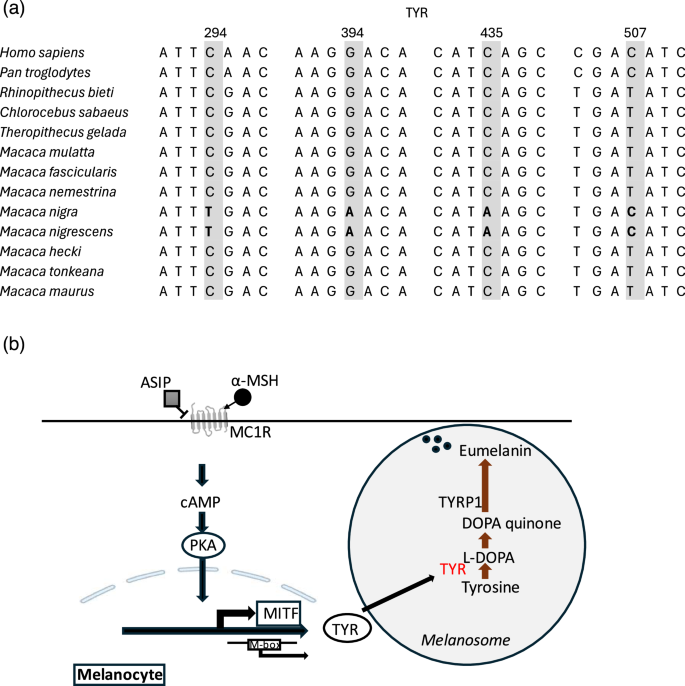
Divergent sites in TYR and its role in the melanogenesis pathway. a Highly divergent SNPs (top 5% Fst) located in the first exon of TYR across primate species. b Schematic graph of TYR’s role in the melanogenesis pathway
Population differentiation analysis focusing on the first exon of TYP revealed that low Fst (0.12) and Dxy (0.0014) between M. nigra and M. nigrescens, contrasting with significantly higher Fst (average 0.89) and Dxy (average 0.0054) values between M. nigra and other Sulawesi macaque species (Table S4). Additional pigmentation-related genes, such as ASIP (Agouti-signaling protein) and HPS5 (Hermansky-pudlak syndrome 5), also exhibited missense variants. In M. nigrescens, a missense variant (E27G) in ASIP, which modulates melanin synthesis, was detected at a highly conserved site. Furthermore, a missense variant (S669L) in HPS5, involved in melanosome trafficking, was identified in M. nigrescens and at low frequencies in M. nigra. These variants may influence pigmentation by altering melanin production or transport mechanisms.
Species-specific genetic marker
We next focused on fixed SNPs, defined as loci with an Fst value of 1 in each paired comparison. Each pairwise comparison contained numerous fixed SNPs, distributed across all chromosomes (Figure S1), with proportions ranging from 0.38 to 1.20% of the total SNPs identified. Although a human exome capture kit was used, a notable fraction of fixed SNPs were located outside annotated coding regions. Among the fixed SNPs, 41.18–45.00% were found in exonic regions. Notably, we identified significantly more fixed SNPs in the NgNc and NcH comparisons than in the other two pairwise comparisons.
For the NgNc comparison, we identified 2,598 fixed SNPs, of which 1,114 SNPs were located within the exons of 705 genes, comprising 306 missense SNPs. Similarly, in the NcH comparison, 2,788 fixed SNPs were found, with 1,226 exonic SNPs spanning 861 genes, including 347 missense SNPs. In the HT comparison, 1,870 fixed SNPs were detected, with 770 exonic SNPs spanning 559 genes, including 258 missense SNPs. Finally, the TM comparison yielded 1,580 fixed SNPs, with 711 exonic SNPs across 516 genes, comprising 225 missense SNPs.
Genes with fixed SNPs across all four comparisons
We identified 11 genes containing fixed SNPs in all four comparisons, suggesting these genes might serve as species markers among Sulawesi macaques (Figure S2). Upon annotating the SNPs positions in these 11 genes, we identified a total of 54 fixed SNPs located on chromosome 2, 4, 6, 14, 16, 20, and X (Table S5). Among these, three variants were located in non-protein coding regions (5’ and 3’ UTR regions) in B4GAT1 (Beta-1,4-Glucuronyltransferase 1), KDM2A (Lysine Demethylase 2 A), and NHS (Nance-Horan Syndrome Protein) genes. Additionally, we detected a 9 bp deletion (GAFCAFCCC/EQP) variant in ZFHX3 (Zinc Finger Homeobox 3) gene specific to M. maurus.
Given the hypothesis that missense variants are rare but can have significant functional consequences, we further concentrated on genes containing fixed missense variants to assess their potential impact on functional divergence among species. The genes analyzed included TGM4 (Transglutaminase 4, missense SNPs, N = 2), MC1R (Melanocortin 1 Receptor, N = 4), NHS (N = 2), BCOR (BCL6 Corepressor, N = 1), and RTL9 (Retrotransposon Gag Like 9, N = 4). TGM4, known for its role in male reproduction, harbored two missense variants (V168M, Y176H), both of which were predicted to have neutral effects based on PROVEAN analysis. Three genes, BCOR, NHS and RTL9, were in X chromosome and contained missense variants. BCOR encodes an epigenetic regulator involved in cell differentiation and body structure development. A missense variant, S613T, which involves a substitution of serine to threonine in exon 4 of BCOR, appears to have a neutral effect on protein function.
NHS, which encodes a Nance-Horan syndrome protein, may play a role in the development of eyes, teeth, and brain. We identified two missense variants located in exon 6 of NHS: A854T (substituting a hydrophobic residue with a polar residue) and V1294A (replacing valine with alanine). Both variants are predicted to have a neutral effect on protein function. RTL9 (Retrotransposon gag-like protein 9, also referred to as RGAG1) is a neofunctionalized retrotransposon primarily expressed in the human testis, although its specific function remains unclear. We detected four missense variants in RTL9 (A197D, S654A, T1015I and G1077C), all located in the first exon. Notably, the T1015I variant may potentially alter functional characteristics, although further experimental validation is needed.
MC1R encodes a receptor crucial for regulating the type and density of melanin synthesis. Four missense variants were identified in MC1R. The I293V variant appears to have a neutral effect, whereas the substitutions in conservative sites (H153P, C267Y, and E304G) are likely to induce significantly functional changes. These missense SNPs in MC1R could alter the functional properties of the receptor, potentially affecting melanin deposition in the macaques’ coat.