This study provided a comprehensive analysis of the genetic landscape of IRD in an Iranian cohort (N = 111), revealing significant insights specific to this population. The diagnostic yield was 59%, with 94% of the identified cases demonstrating AR homozygous inheritance. This underscores the significant impact of consanguinity on the genetic architecture of IRD in this population. Notably, our study identified 14 previously unreported causal genetic variants across 12 genes, thereby expanding the current spectrum of IRD genetics and contributing to the global knowledge of IRD genetic diversity.
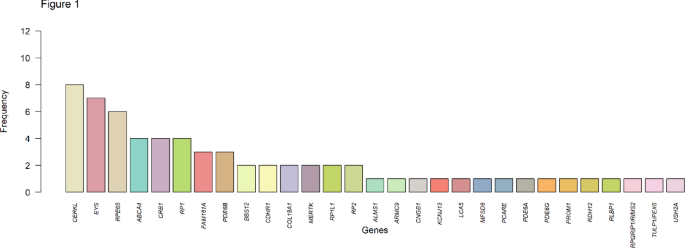
WES was employed using the vision disorders exome sequencing panel from the clinical genetics department at Erasmus MC, University Medical Center, which is specifically designed to capture a wide array of vision-disorder-related genes15. This approach facilitated an in-depth analysis of genetic variations within this Iranian cohort. WES identified causative genetic variants in 59% of the patients, in 31 distinct genes. The variants discovered included a mix of frameshift, nonsense, missense, and splice site mutations, with frameshift mutations being the most prevalent (36%). The most frequently mutated genes were CERKL, EYS, RPE65, RP1, CRB1 and ABCA4, collectively accounting for over half of the detected variants. Particularly, variants in ABCA4, RPE65, and CRB1 have been previously recognized as important causal genes in the Iranian population16,17. These findings highlight the importance of these genes in pathogenesis of IRD in this population. Furthermore, a comparison of the causal genes identified in this study with those in other global populations reveals that these genes are consistently among the most frequently implicated in IRD pathology18,19. Interestingly, variants in USH2A were observed to be less prevalent in our study population compared to other IRD populations20,21,22.
IRD follow various inheritance patterns. For non-syndromic RP, 15-25% of cases demonstrate AD inheritance, 5-20% AR inheritance and 5–15% are X-linked23. In 62 of the 66 solved cases (94%), the causal variants were homozygous with an AR inheritance pattern, reflecting the high levels of consanguinity in our Iranian cohort. Therefore, genetic analyses in this population should prioritize the filtering of homozygous variants, for which WES is particularly well suited24.
The diagnostic yield in this study is comparable to other studies utilizing WES analysis in specific IRD populations (51–57%)17,20,25. VUS were identified in 35 patients (32%). Due to limited opportunities for further family segregation analysis or additional clinical investigations, the genetic cause could not be definitively established in these cases. Consequently, the diagnostic yield in our population may, in fact, be higher.
Among the six unsolved cases, potential explanations include pathogenic variants in non-coding regions, structural variants undetected by WES, or involvement of unknown disease-associated genes.
This study also emphasized genetic variations among different ethnic groups within Iran. Variants in the CERKL gene were more frequently observed among individuals of Turkish descent, while RPE65 variants were predominant among individuals of Kurdish descent and variants in EYS were most observed in individuals from Fars. These findings highlight the varying prevalence of certain variants among different ethnic groups and emphasize the importance of personalized genetic counseling and diagnostic strategies.
Fourteen novel variants (26%) were identified, this underscores the unique genetic profile of this Iranian population. Furthermore several genetic variants identified in our study, have been recently reported for the first time in various populations. In the CERKL gene, variant c.560_568del9, has been newly documented in an Iranian family26. Similarly, the c.847 C > T variant, was recently discovered in a Pakistani family27. In the EYS gene, variant c.32dupT was first reported in 2017 in an proband from East Indian/Iranian descent, and the c.490 C > T in 2019 in a Chinese family28,29. The c.731G > A variant in the RPE65 gene, classified as a VUS, has been previously reported in a Turkish population30. Additionally, the c.1879T > C variant in the CRB1 gene was recently identified in an Iranian population31. The c.2927delT variant in the ABCA4 gene in an Iranian family, and variant c.2824_2831del8 in the COL18A1 gene in an English cohort, have been reported only once32,33. Lastly, the c.1256G > A variant in the TULP1 gene have been previously reported in one single individual from Saudi-Arabia34. Furthermore several variants are specifically reported in Middle-Eastern and Asian populations35,36,37,38,39. Our findings reinforce the recently reported variants and facilitate the identification of the genetic causes of IRD in these populations in the future.
The novel variants identified in this study were evaluated in gnomAD (v.4.1.0, https://gnomad.broadinstitute.org/), to determine their population frequency. Given the high rate of consanguinity in the Iranian population, some variants may show an elevated frequency within our in-house control cohort. This factor was considered in the interpretation of variant significance.
This study contributes to creating a more inclusive genetic landscape of IRD. To develop a truly representative overview of the genetic landscape of IRD, it is imperative to include all countries in genetic analyses. This inclusivity will enhance our understanding of genetic diversity, reduce health disparities, and improve the development and accessibility of gene-based therapies globally9. As the field of genetics continues to advance, a concerted effort to integrate diverse populations will ensure that the benefits of genetic research are equitably distributed. Ultimately, contributing to better health outcomes for all individuals affected by IRD. Comparing our Iranian cohort with a Dutch IRD cohort revealed both similarities and differences in the most prevalent causal genes40. While certain genes were common to both populations, others were unique to the Iranian cohort, representing the genetic diversity and population-specific factors influencing IRD prevalence and mutation patterns. In patient care, an awareness of patients’ diverse migration backgrounds is thus essential, as ancestry can significantly influence the genetic factors underlying inherited conditions. By incorporating ancestry into diagnostic considerations, we can improve diagnostic accuracy and provide more tailored patient care, highlighting the value of cultural and genetic diversity in medical practice and research.
Furthermore accurate genetic diagnosis is crucial for effective IRD management and genetic counselling for family and patients, especially when clinical symptoms overlap. In this study, six patients were initially diagnosed with non-syndromic IRD; however, genetic analyses revealed syndromic IRD, indicating the broader implications of some IRD-related variants. Unfortunately, the syndromic diagnosis in these patients could not be clinically confirmed, as prior systemic evaluations were not available. Further clinical assessment would be necessary to fully characterize the systemic manifestations of these cases. Nonetheless, these findings underscore the critical role of genetic testing in identifying syndromic cases that might otherwise go unrecognized.
Genetic diagnostics for IRD in Iran currently cover only a small portion of the affected population. This is largely due to limited local resources and infrastructure, which often necessitate that genetic testing needs to be conducted abroad. For instance, in the first national IRD registry in Iran, only about 20% of registered individuals have undergone genetic testing41. This indicates a gap in access to genetic diagnostics within the country, highlighting the need for further development of local genetic testing capabilities.
To enhance clinical advice informed by genetic outcomes, it is essential to integrate clinical measurements with genetic analyses. In this study, we faced limitations in accessing all clinical data, partly due to variations in diagnostic procedures. However, in clinical practice, the integration of ophthalmic examinations with the genetic outcome is relevant.
In conclusion this study provides a detailed analysis of IRD in an Iranian cohort, highlighting the prevalence of autosomal recessive inheritance due to high grade of consanguinity. The discovery of novel genetic variants expands the understanding of IRD genetic diversity and highlights the importance of including diverse populations in genetic studies to enhance diagnosis, treatment, and equitable access to gene-based therapies globally.