Identification of the gene cluster involved in siderophore biosynthesis
Based on the complete genome of T. maritimum SP9.1 (GenBank: CP170088.1)9 an in silico search was performed to identify candidate genes involved in siderophore biosynthesis (Fig. 2; Table 1). This analysis revealed a cluster of six genes encoding functions homologous to T. mesophilum bsbABECDC2D2 genes (showing 74–92% similarity), as well as to the desEFABCD genes of Streptomyces coelicolor (60–71% similarity), which encode desferrioxamine biosynthesis.
Gene content and transcriptional organization related to siderophore biosynthesis in Tenacibaculum maritimum SP9.1. (a) The tenABECDC2D2 genes are represented by colored arrows, while downstream genes are shown in white arrows. “RT” indicates the location of the primers used for the reverse transcription reactions 1 and 2 (RT-1 and RT2), and “PCR” denotes the primer pair used for subsequent gene sequence amplification. The black arrow marks the insertion site of plasmid pPE17 to generate the tenCD disruption mutant. (b) Operon mapping: RT-PCR amplifications obtained from T. maritimum cultures under low-iron (E) and high-iron conditions (F). Numbers on the left indicate molecular size in kb (M). Positive controls (+) correspond to PCRs using chromosomal DNA as a template, while the negative controls (–) are RT-PCR reactions performed without reverse transcriptase. The agarose gel images were cropped for clarity and to conserve space (the original image can be found in the Supplementary Information).
The organization of the T. maritimum siderophore gene cluster differs from that of S. coelicolor, as it includes a major facilitator superfamily (MFS) exporter between desB and desC and features a duplication/fusion of the desCD genes (Fig. 2; Table 1). The genes identified in T. maritimum SP9.1 were designated as tenABECDC2D2, based on their predicted functions (Table 1).
Table 1 Gene cluster encoding the HSC-based siderophore in T. maritimum SP9.1 and its homologs in T. mesophilum 13764 (NCBI: txid104268) and Streptomyces coelicolor strain A3(2) / M145 (NCBI: txid100226). Homologous ORFs shared between clusters are presented with their percentage identities and similarities at the amino acid sequence level.
This T. maritimum gene cluster likely encodes the following functions: lysine decarboxylase (tenA), lysine 6-monooxygenase (tenB), major facilitator transporter (tenE), desferrioxamine E synthases (tenCD), transferase (tenC2), and desferrioxamine D synthase (tenD2). Notably, tenCD encode acyl-CoA acyltransferase and ligase domains, with each domain sharing 67% and 65% nucleotide homology with tenC2 and tenD2, respectively. The relatively low homology between tenCD and its counterparts, tenC2 and tenD2, suggests that they are unlikely to have originated from a duplication event.
Interestingly, four additional genes are located downstream of tenD2 on the same strand (Fig. 2A). These genes are also conserved in T. mesophilum, demonstrating synteny (Fig. 2; Table 1). They likely encode a nitroreductase (hp1), N-acetyltransferase (hp2), lipase/esterase (hp3), and sterol desaturase (hp4). The 10 genes, including tenABECDC2D2 and hp1-4, are flanked by fldA (flavodoxin) and araC (an AraC-like transcriptional regulator), which are encoded on the opposite strand (Fig. 2A). This genomic region is nearly identical in T. maritimum SP9.1 and the type strain NCIMB 2154T, sharing 99.5% nucleotide identity. Although the functions encoded by hp1-4 do not seem to be directly related to siderophore synthesis, their gene arrangement suggests that they could be co-transcribed as part of a polycistronic mRNA along with the ten siderophore gene cluster.
Insertional inactivation of the ten gene cluster significantly reduces siderophore production
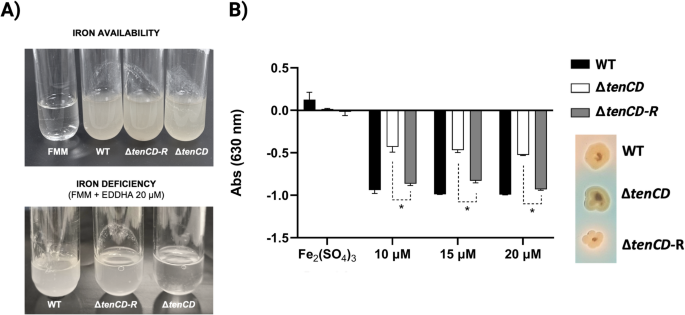
Both the wild-type and its derivative insertional mutant ΔtenCD (tenCD::pPE17), exhibited nearly identical growth under high-iron conditions (Fig. 3A). However, under iron-restricted conditions (FMM medium supplemented with 10, 15, or 20 µM EDDHA), the ΔtenCD mutant showed reduced growth (Fig. 3A) and diminished siderophore production, as measured by the CAS assay (Fig. 3B), compared to the wild-type strain (Fig. S1).
Notably, the insertional mutant was highly unstable, spontaneously losing the plasmid when cultivated with and without antibiotic selection, leading to the formation of a revertant strain (ΔtenCD-R). This revertant strain, which regained the full-length tenCD gene, exhibited a phenotype indistinguishable from the wild-type strain in terms of both growth and siderophore production (Fig. 3). The spontaneous loss of the plasmid, despite the application of selective pressure, prevented us from obtaining supernatants for siderophore analysis. Although the insertional inactivation of tenCD could have polar effects on downstream genes and was not permanent, our results strongly suggest that the ten gene cluster, including tenCD, is conserved in T. maritimum and that its disruption impairs siderophore production.
(A) Growth assay of T. maritimum NCIMB 2154T wild-type (WT) and its derivative strains (ΔtenCD and ΔtenCD-R) under iron-suficient and iron-deficient conditions (B) Siderophore production (CAS assay) of T. maritimum NCIMB 2154T wild-type (WT) and its derivative strains (ΔtenCD and ΔtenCD-R). Three independent experiments were performed for each condition, and standard deviations are shown as error bars. Asterisks indicate statistically significant differences among strains (p 25. Picture to the right illustrates phenotype of each strain grown on CAS-Agar.
T. maritimum isolates exhibit homogeneous growth and siderophore production, consistently carrying the tenABECDC2D2hp1-4 siderophore gene cluster
To investigate the distribution of siderophore biosynthetic genes in T. maritimum, a collection of 64 strains (including SP9.1) was analyzed. These strains, isolated from disease outbreaks in various locations and hosts and representing all four serotypes (O1-O4), were subjected to PCR screening to detect the five genes encoding biosynthetic functions (tenA, tenB, tenCD, tenC2, and tenD2). All 64 strains tested positive for amplification of these genes, indicating the presence of the ten siderophore biosynthetic gene cluster (Table S1). These findings reinforce the notion that T. maritimum universally harbors a complete and conserved ten gene cluster.
To further assess growth capacity and siderophore production, the strains were cultivated in FMM medium, under iron-replete conditions and progressively lower iron availability, achieved by supplementing the medium with the iron chelator EDDHA at concentrations of 10, 15, and 20 µM. All strains exhibited phenotypes nearly identical to T. maritimum SP9.1 and were capable of growing under iron-restricted conditions up to 20 µM EDDHA. Notably, all tested isolates produced siderophores, as evidenced by a CAS-positive phenotype under iron-limited conditions. These results strongly suggest that the siderophore biosynthetic gene cluster is highly conserved across T. maritimum isolates, demonstrating homogeneity in both siderophore production, as measured by the CAS assay, and growth behavior under iron-deficient conditions.
The siderophore gene cluster is transcribed as a polycistronic mRNA
To study the transcriptional organization of the T. maritimum siderophore gene cluster (the tenABECDC2D2 operon) and determine whether it is co-transcribed with the downstream genes, two reverse transcription reactions were performed using primers targeting the 3′ end of tenD2 and hp4 (Fig. 2B). The resulting cDNA was then subjected to PCR with primers targeting tenA (PCR1), tenB (PCR2), tenE (PCR3), and tenD2 (PCR4) (Fig. 2B). Both cDNAs from RT-1 and RT-2 showed positive PCR amplification for PCR1. Furthermore, the intensity of the amplicons was higher when bacteria were cultivated under iron-starvation conditions (E) compared to iron-replete conditions (F). These results suggest that the siderophore gene cluster is transcribed from a promoter located immediately upstream of tenA, within the fldA-tenA intergenic region (Fig. 2A). The possibility of an alternative promoter in the tenB-tenE intergenic region cannot be ruled out, as this region spans 304 bp, and PCR3 amplicons showed increased intensity under both iron-starvation and iron-replete conditions. Taken together, these findings confirm that the siderophore gene cluster is transcribed as a polycistronic mRNA, spanning from tenA to hp4.
Identification of siderophores produced by T. maritimum SP9.1
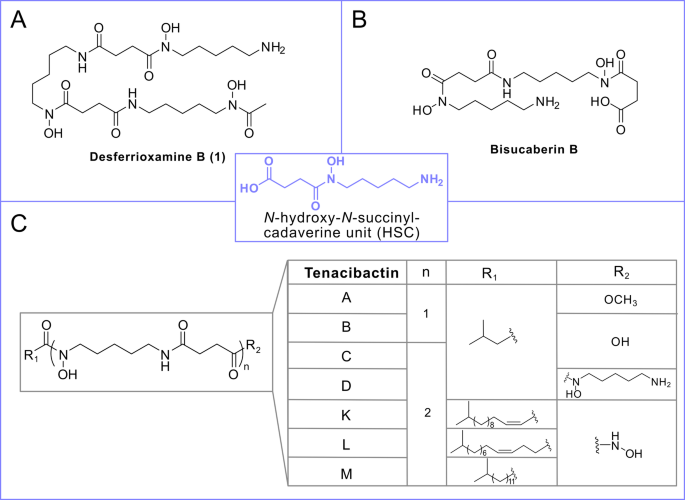
In a parallel study we employed a novel analytical methodology, XAD-LC/MS-FBMN-IMS21 to investigate the siderome of T. maritimum LSP9.1, a strain isolated from a tenacibaculosis outbreak in cultured Solea senegalensis. Chemical analysis of this strain revealed three families of hydroxamate siderophores, including three known desferrioxamine derivatives and 17 novel putative acyl-desferrioxamine-like structures21. In the present study, we confirm that the same three families of hydroxamate siderophores are also produced by a distinct strain, T. maritimum SP9.1, isolated from Salmo salar. LC-HRESI-MS/MS analysis of extracts from cell-free supernatants and cell pellets detected the presence of 20 putative desferrioxamine-like siderophores. Structural annotation based on MS/MS data revealed a common core structure comprising N-hydroxy-N-acyl-cadaverine (HAC) and N-hydroxy-N-succinyl-cadaverine (HSC) units. Structural variations among siderophores were found at both terminal ends. At one terminal end, variations included acetylated cadaverine, a hydroxamic group or succinic acid, allowing classification into three distinct families (Fig. 4). Differences within each family depend on the length and degree of unsaturation of the fatty acid tail in the N-hydroxy-N-acyl-cadaverine unit at the other terminal end.
The first group, N-acetylated-cadaverine desferrioxamines, includes the well-known desferrioxamine D1 (2) and two previously identified siderophores from Streptomyces spp.: C4 acyl DFO-D (3) and C5 acyl DFO-D (4)20,26 along with five newly proposed structures, compounds 5–9. This group features analogues of desferrioxamine D1 (DFO-D) with various acyl chains at the N-hydroxy-N-acyl-cadaverine unit.
The second group of siderophores has a hydroxamic acid terminus, similar to tenacibactins K, L, and M (Fig. 1C)19. MS/MS fragmentation analysis identified seven new tenacibactin-like siderophores, designated as compounds 10–16 (Table 2; Fig. 4).
The final family of siderophores is characterized by a succinic acid terminus as in tenacibactins B, C, and bisucaberin B (Fig. 1). Within this group, two new structures, compounds 20 and 21, were proposed, while other three (compounds 17–19) were detected but could not be structurally resolved by MS/MS data. Interestingly, although bisucaberin B is reported as the siderophore produced by T. mesophilum, a species closely related to T. maritimum, this compound was not detected in the T. maritimum siderome.
Table 2 Potential siderophores detected in the cell pellets and cell-free supernatants of Tenacibaculum maritimum SP9.1.Fig. 4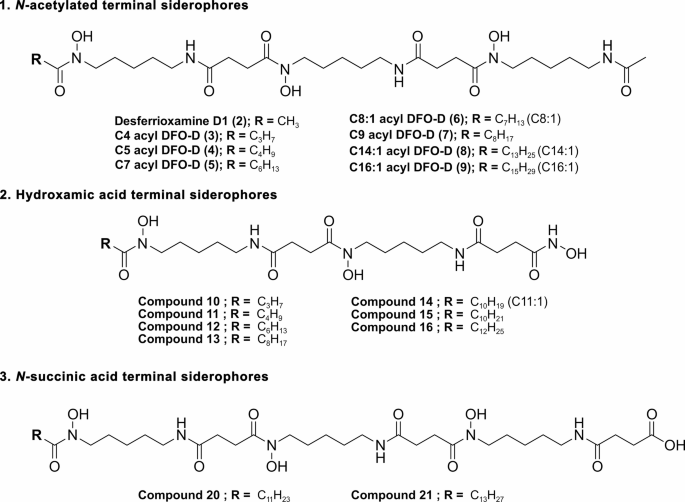
Proposed structures of the metallophores detected through the XAD-LC/MS-FBMN-IMS methodology correspond to three distinct families: (1) acetylated desferrioxamine-like compounds, (2) hydroxamic acid-terminal compounds, and (3) succinic acid-terminal compounds, as described in Ageitos et al.21. The precise positions and configurations of the double bonds, as well as the detailed structure of the acyl chain (whether linear or branched), have yet to be elucidated.
As described above, it is noteworthy that all T. maritimum strains tested harbor the ten gene cluster. Therefore, it is reasonable to infer that the siderophore families identified in this study are a conserved trait of the species.