Inclusion criteria and ethical considerations
Between 7.5 and 15 ml of patient blood in EDTA vacutainers was collected upon written informed patient consent. All specimens were obtained at the University Hospital Basel under ethical approval from the Ethics Committee Northwestern and Central Switzerland (EKNZ), in accordance with the Declaration of Helsinki (protocols EKNZ BASEC 2016-00067, EKNZ 2014-329 and EK 321/10). The patients did not receive any participant compensation. The clinical characteristics of interrogated patients with cancer are included in Supplementary Table 1. All mouse experiments were carried out according to institutional and cantonal guidelines (mouse protocol 33688, approved by the cantonal veterinary office of Zurich).
Cell culture
MDA-MB-231 lung metastatic variant 2 (LM2) human breast cancer cells (obtained from J. Massagué, Memorial Sloan Kettering Cancer Center) were grown in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (Gibco, 11330032) supplemented with 10% FBS (Gibco, A5256801) and 1× Antibiotic–Antimycotic (Gibco, 15240062) in a humidified incubator at 37 °C with 20% O2 and 5% CO2. Human CTC-derived BR16 cells were generated as previously described28 from a patient with hormone receptor-positive breast cancer at the University Hospital Basel and propagated as suspension cultures in a humidified incubator at 37 °C with 5% O2 and 5% CO2. LM2 and BR16 cells were labeled with a GFP-luciferase construct through lentiviral transduction. Cell lines do not belong to the list of commonly misidentified cell lines (International Cell Line Authentication Committee) and were confirmed negative for common contaminating microorganisms, including mycoplasma, by an independent laboratory. Cells were not authenticated as authentication is not applicable for the BR16 and LM2 cell lines.
Molecular barcoding of LM2 cells
For barcoding experiments, LM2-GFP-luciferase cells were transduced with the CloneTracker XP 5M Barcode-3′ Library in vector pScribe4M-RFP-Puro (Cellecta, BCXP5M3RP-1S-V), containing 4.8 million unique barcode combinations packaged into lentiviral particles. Cells were transduced at a multiplicity of infection below 0.1 to obtain a high proportion of cells with a single, unique barcode integration. Seventy-two hours after lentiviral transduction, barcoded cells were selected based on red fluorescent protein (RFP) signal via fluorescence-activated cell sorting and immediately processed for transplantation into mice.
Mouse experiments
All mouse experiments were carried out according to institutional and cantonal guidelines (mouse protocol number 33688, approved by the cantonal veterinary office of Zurich). Experimental endpoints specified in our approved license, comprising tumor-related factors, as well as behavioral and appearance-related factors, were closely monitored. The tumor size never exceeded the maximum permitted limit of 2,800 mm3. Replacement, reduction and refinement (3R) principles were considered and complied with throughout all experiments. Female NSG mice were purchased from The Jackson Laboratory and kept in pathogen-free conditions in a controlled environment with a room temperature maintained at 22 ± 2 °C and relative humidity at 55 ± 10%, according to institutional guidelines. Animals were kept under a standard 12-h light/12-h dark photoperiod.
Orthotopic breast cancer lesions were generated in eight-week-old to ten-week-old NSG females upon injection of 106 LM2-GFP-luciferase or BR16-GFP-luciferase cells into the mammary fat pad. In both cases, breast cancer cells were inoculated in 100 μl of 50% Cultrex Reduced Growth Factor Basement Membrane Extract, Type 2, PathClear (BME, R&D Biosystems, 3533-010-02) in Dulbecco’s PBS (Gibco, 14190144). Terminal blood draws through cardiac puncture for CTC analysis were performed after four to five weeks for LM2-NSG and five months for Br16-CDX-NSG models.
For barcoding experiments, barcoded LM2-GFP-luciferase cells were inoculated in a 1:1 mix of BME and DPBS at densities corresponding to 102, 103, 104 and 5 × 104 cells in 100 μl. Orthotopic breast cancer lesions of varying barcode complexities were induced upon injection of 100 μl of generated cell suspensions into the mammary fat pad of female NSG mice. All animals were injected and sacrificed synchronically to prevent variability due to circadian fluctuations. No animals or data points were excluded from the analysis.
CTC capture and immunofluorescence staining
Patient-derived CTCs were captured from unprocessed peripheral blood samples using the FDA-approved microfluidic device Parsortix (ANGLE) equipped with Cell Separation Cassettes (ANGLE, GEN3D6.5). In-cassette staining was performed with antibodies against EpCAM-AF488 (1:50; Cell Signaling Technology, CST5198), HER2-AF488 (1:50; BioLegend, 324410), EGFR-FITC (1:25; GeneTex, GTX11400) and CD45-BV605 (1:25; BioLegend, 304042). Mouse-derived CTCs were captured from 0.8 to 1.2 ml of blood in EDTA tubes (Sarstedt, 41.3395.005) collected through cardiac heart puncture using the Parsortix Cell Separation System as described above and identified based on GFP expression due to stable expression of a GFP-Luciferase reporter. Anti-CD45 staining was carried out to identify CD45-positive cells within the cassette. Microscopic images were processed using the Fiji image processing software (v2.14.0).
For barcoding experiments, CTCs were identified based on the expression of both GFP and RFP, due to stable RFP expression from the integrated barcode cassette. All CTCs were released from Cell Separation Cassettes in reversed flow direction with Dulbecco’s PBS onto ultra-low-attachment plates (Corning, 3471-COR) for downstream procedures.
Micromanipulation of CTCs and CTC clusters
Whenever possible, CTC clusters for exome sequencing were mechanically dissociated through gentle micromanipulation (CellCelector, ALS). Individual cells from dissociated CTC clusters, intact CTC clusters and single CTCs were aspirated using the automated single-cell picking system CellCelector (ALS) and deposited into individual PCR tubes (Axygen, 21-032-501) containing 2.5 μl RLT Plus lysis buffer (Qiagen, 1053393) and 1 U μl−1 SUPERase In RNase Inhibitor (Invitrogen, AM2694). Samples were immediately frozen on dry ice and kept at −80 °C until further processing.
For barcoding experiments, intact CTC clusters were picked as described above and deposited into individual PCR tubes containing 1 μl of oligo-dT primer, 1 μl of dNTP mix and 2.3 μl of cell lysis buffer (0.2% (vol/vol) Triton X-100 (Sigma-Aldrich, X-100) and 2 U μl−1 SUPERase In RNase Inhibitor). Samples were immediately frozen on dry ice and transferred to −80 °C until further processing. All pre-PCR steps were carried out in a PCR cabinet with laminar air flow to reduce environmental contamination.
Primary tumor processing
Barcoded primary tumors were surgically resected from mice after terminal blood sampling, transferred to 50 ml screw-cap tubes (Sarstedt, 62.547.254) containing precooled CO2-Independent Medium (Gibco, 18045-088) and stored on ice until further processing. Subsequently, tumor tissue was transferred to Lysing Matrix S tubes (MP Biomedicals, 116925500) and homogenized on a Precellys 24 tissue homogenizer (Bertin Technologies) for 2 × 20 s at 5,500 rpm. Homogenized tumor tissue was transferred to 50 ml screw-cap tubes and lysed in 18 ml of tissue lysis buffer (40 mM TRIS pH 8, 1% SDS and 50 mM EDTA) supplemented with 100 μl Proteinase K (Qiagen, 19133) with constant shaking at 55 °C overnight. The next day, 100 μl of 100 mg ml−1 RNAse A (Qiagen, 19101) was added; tubes were thoroughly mixed through inversion and incubated with constant shaking for 30 min at 37 °C. After that, tubes were immediately chilled on ice before the addition of 9 ml precooled 7.5 M ammonium acetate solution (Sigma-Aldrich, A2706), followed by thorough mixing through inversion of tubes and rigorous vortexing for 1 min at full speed to reduce the molecular weight of DNA. Subsequently, the tubes were centrifuged at 4,400g for 10 min at 4 °C to precipitate salts and proteins. DNA was recovered from the supernatant by decanting on top of 20 ml of 100% isopropanol in a fresh 50 ml tube. Tubes were mixed by inversion 50 times and centrifuged at 4,400g for 15 min at 4 °C to pellet DNA. Supernatants were discarded and DNA pellets were purified twice with precooled 70% ethanol. Ethanol was removed and DNA pellets were dissolved in TE buffer (Invitrogen, 12090015) over constant agitation. Dissolved DNA samples were sheared in 1 ml AFA Fiber milliTUBEs (Covaris, 520135) on a LE220-plus instrument (Covaris) for 60 s with 200 cycles per burst, a duty factor of 10% and a peak incident power of 450 to reduce the molecular weight of DNA and increase PCR efficiency.
Exome sequencing
Exome sequencing of CTC samples was performed based on the previously published G&T-seq protocol29. Genomes and transcriptomes of lysed cells were separated, and genomes were amplified using the GenomiPhi V3 Ready-To-Go DNA Amplification Kit (Cytiva, 25-6601-97). Libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina, FC-131-1096); exomes were enriched using the SureSelect XT Human All Exon v6 + Cosmic Kit (Agilent Technologies, 5190-9308) and sequenced on a HiSeq 2500 instrument (Illumina) in 100 bp paired-end mode.
Exome sequencing analysis
Paired-end reads were aligned to the GRCh38 human reference using BWA-mem algorithm (v0.7.15)30 and sorted using SAMtools (v.1.7)31. Xenograft samples were additionally aligned to the GRCm38 mouse reference genome and assigned to either human or mouse using Disambiguate (v1.0.0)32. Reads identified as mouse were removed from subsequent analysis. Deduplication of reads was performed on a per-sample basis using Picard MarkDuplicates (v.2.9.2), and local realignment was performed using the Genome Analysis Toolkit IndelRealigner (v.3.7.0)33 at the sample and donor level to improve alignment accuracy around indels. Quality control as well as coverage and exome enrichment statistics were generated using FastQC (v.0.11.8), CollectHsMetrics from Picard suite (v.2.9.0) and QualiMap (v.2.2.1)34 and visualized using MultiQC (v.0.8)35. Mpileup files were generated with SAMtools (parameters: -q 40 -Q 30) at donor level, and variants were called using SCIΦ on all samples from the same donor simultaneously.
Genetic variant annotation
The variant annotation and effect prediction tool SnpEff (v.5.2a)36 was used to classify observed genetic variants by putative impact on protein functionality, using default parameters and variant calling format files as input. The Cancer Genome Interpreter web tool was used to analyse genetic variants by their predicted oncogenic capacity37.
Barcode sequencing
Amplified cDNA was obtained for individual CTC cluster samples following the previously published SmartSeq2 protocol38. Barcode loci were amplified from purified cDNA (CTC cluster samples) or sheared gDNA (primary tumor samples) using the KAPA HiFi HotStart ReadyMix (Kapa Biosystems, KK2602) supplemented with 5% (vol/vol) DMSO and a set of equimolar pools of staggered primers flanking the barcode locus (final concentration of pools = 300 nM), following cycling conditions according to the manufacturer’s recommendations with a primer annealing temperature of 63.5 °C. Barcode amplicon samples were then submitted to a second PCR step to introduce unique dual indexes, sequencing primer-binding sites and Illumina adapter sequences P5 and P7. All primers used are listed in Supplementary Table 3. All PCR steps were performed in a T100 Thermal Cycler (Bio-Rad). Final amplicons were purified using AMPure XP Beads (Beckman Coulter, A63881) and sequenced on an Illumina NovaSeq instrument in 150-base-pair paired-end mode to generate files in FASTQ format.
Barcode analysis
Reads in FASTQ files were aligned to barcode reference sequences using bowtie2 (v2.5.1; parameters: –local –score-min L,130,0), considering only reads aligning in full length without mismatch. Resulting SAM files were sorted using Samtools sort (v1.16.1), and the number of read segments mapped to each barcode reference sequence was counted using Samtools idxstats (v1.16.1). The resulting barcode count files were processed in R (v4.2.3, R Foundation for Statistical Computing) for secondary analyses. Taking into consideration an expected single barcode integration event per cell, samples were removed from downstream analyses when the smallest number of distinct barcodes accumulating 90% of total aligned reads was higher than the expected number of cells in the sample, indicating profound background noise contribution as seen in negative control samples. CTC cluster samples were classified as monoclonal or oligoclonal based on the detected barcode distribution, taking into consideration the read count of the most abundant barcode relative to the second most abundant barcode and the number of cells in the corresponding CTC cluster sample. A CTC cluster was determined to be monoclonal whenever the read proportion of the most dominant barcode exceeded the read proportion of the second most abundant barcode multiplied by the number of cells in the cluster. Otherwise, the CTC cluster was determined to be oligoclonal.
Statistics and reproducibility
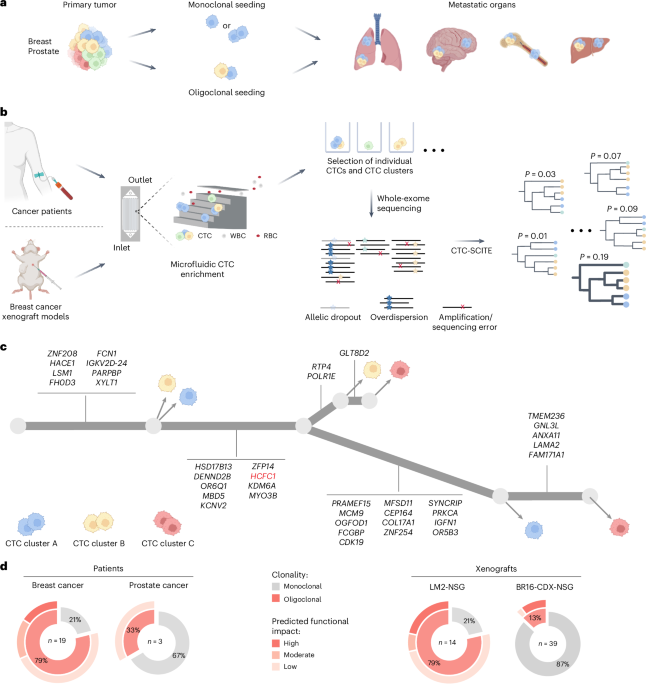
Statistical testing and visualizations were conducted in R (v4.2.3, R Foundation for Statistical Computing). Graphical illustrations in Figs. 1a–c and 2a and Extended Data Figs. 2a and 3b were generated using BioRender.com and Adobe Illustrator. No statistical method was used to predetermine sample size. For mouse experiments, sample sizes were determined in accordance with the 3R principles and consistent with those reported in previous publications13,24. No data were excluded from the analyses. All mice were randomized before experiments and blindly selected before tumor cell injection. Two independent animal experiments were performed, confirming the reproducibility of our findings.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.